Search Count: 457
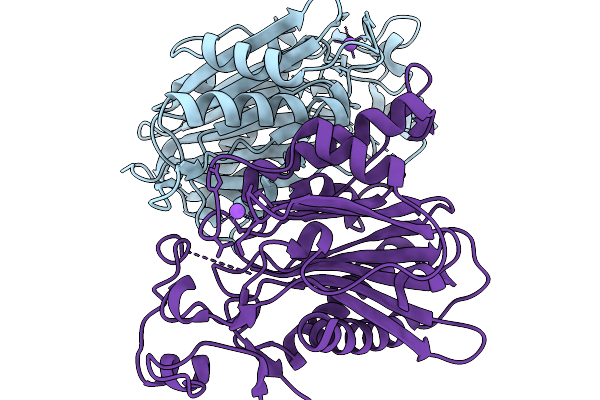 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE |
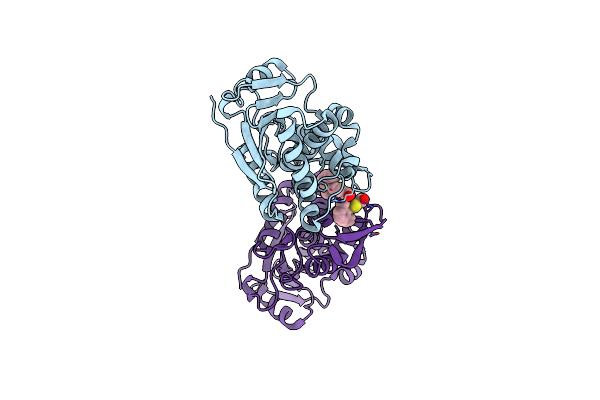 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH4, CL |
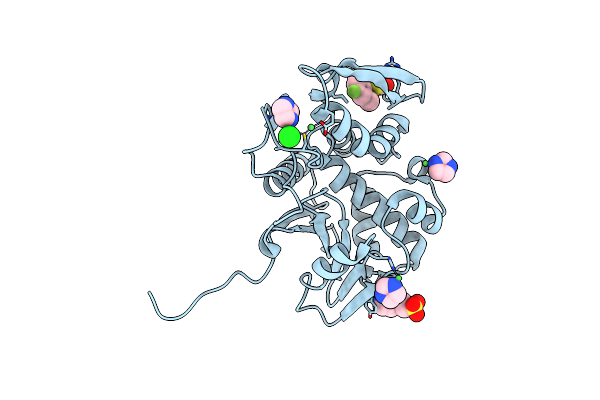 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH3, IMD, EPE, NI, CL |
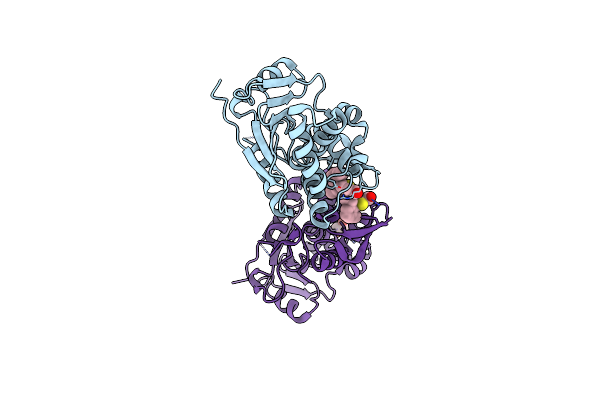 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH2, EDO |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
 |
Organism: Tgev virulent purdue, Canis lupus familiaris
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: ISOMERASE Ligands: NAG |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthse Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Formyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, FYN, MG, ADP, EDO, PO4, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Coenzyme A, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-10-02 Classification: LYASE Ligands: FYN, TPP, MG, ADP, EDO, SO4, CL, GOL |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-05-01 Classification: TOXIN Ligands: ZJT, 2PE |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-05-01 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: CL, U4T, EDO, 2PE |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-05-01 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: ZXJ, CL, 2PE |
 |
Crystal Structure Of Ricin A Chain Bound With N2-(2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carbonyl)Glycyl-L-Tyrosine
Organism: Ricinus communis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-09-20 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of Ricin A Chain Bound With (2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carbonyl)-L-Tyrosine
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: I8T, SO4 |
 |
Crystal Structure Of Ricin A Chain Bound With (2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carbonyl)-D-Tyrosine
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: I9P, SO4 |
 |
Crystal Structure Of Ricin A Chain Bound With (2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carbonyl)-L-Phenylalanine
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: IEK, SO4 |
 |
Crystal Structure Of Ricin A Chain Bound With (2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carbonyl)-D-Phenylalanine
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: IEW, SO4 |
 |
Crystal Structure Of Ricin A Chain Bound With (S)-2-(2-Amino-4-Oxo-3,4-Dihydropteridine-7-Carboxamido)-3-(4-Fluorophenyl)Propanoic Acid
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: IEZ, SO4 |

