Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
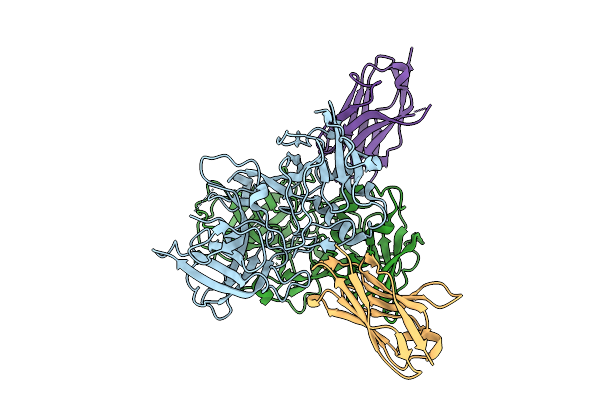 |
Organism: Hare calicivirus, Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN |
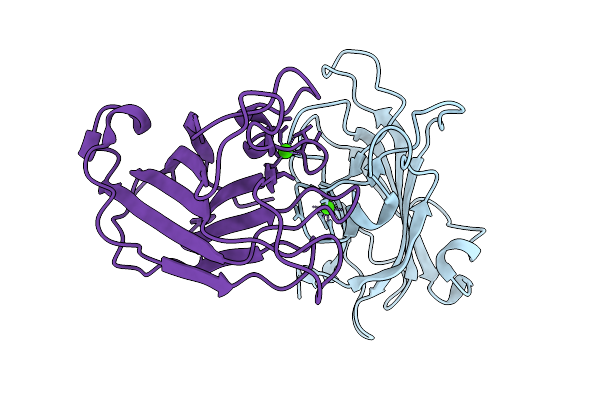 |
Carbohydrate-Binding Module 32 Of Lnbb From Bifidobacterium Bifidum, Ligand Free Form, Multiple Small-Wedge Data Set
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: CA |
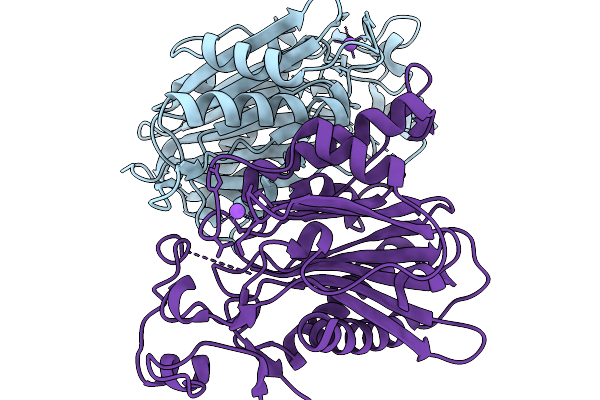 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE |
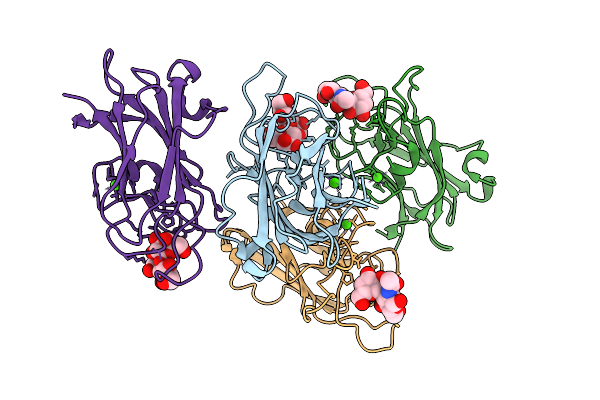 |
Carbohydrate-Binding Module 32 Of Lnbb From Bifidobacterium Bifidum, Lnb Complex
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: SUGAR BINDING PROTEIN Ligands: CA |
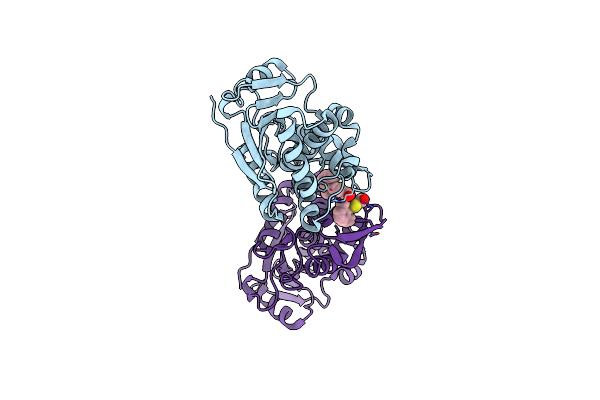 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH4, CL |
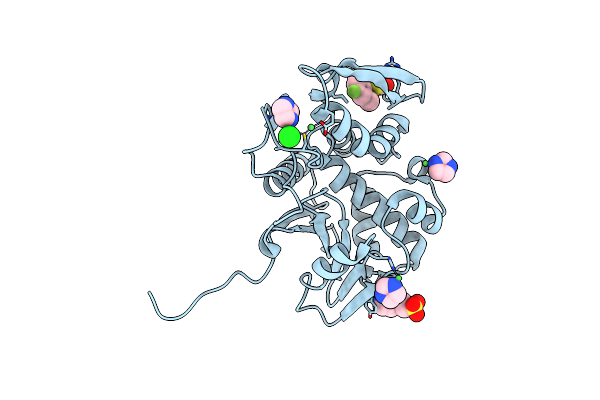 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH3, IMD, EPE, NI, CL |
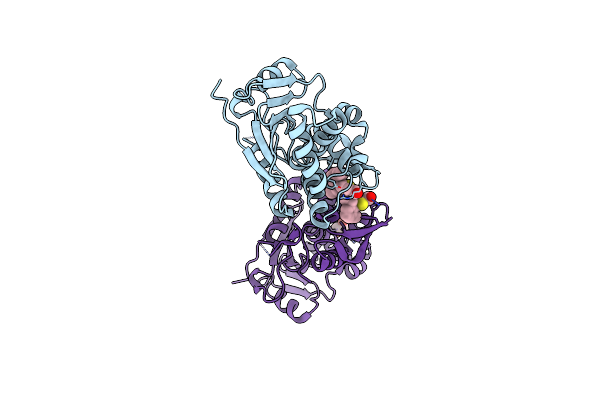 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH2, EDO |
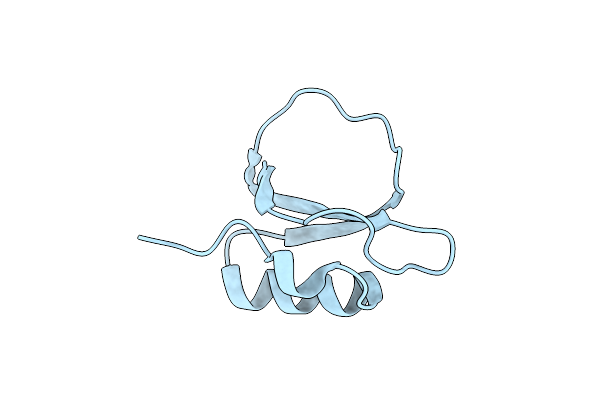 |
Crystal Structure Of A Serine Protease Inhibitor Hpi From Hevea Brasiliensis
Organism: Hevea brasiliensis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-30 Classification: PLANT PROTEIN |
 |
Crystal Structure Of The Dimeric Transaminase Doed From C. Salexigens Dsm 3043.
Organism: Chromohalobacter israelensis dsm 3043
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: GOL, PLP |
 |
Organism: Allochromatium tepidum
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PHOTOSYNTHESIS Ligands: BCL, CA, CRT |
 |
Organism: Allochromatium tepidum
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PHOTOSYNTHESIS Ligands: BCL, CRT |
 |
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-02-19 Classification: TRANSFERASE Ligands: NCO, PGE, GOL, IPA, SAH, CL |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
 |
Organism: Hevea brasiliensis
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: HYDROLASE Ligands: GOL, MG, CL |
 |
Organism: Tgev virulent purdue, Canis lupus familiaris
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: ISOMERASE Ligands: NAG |
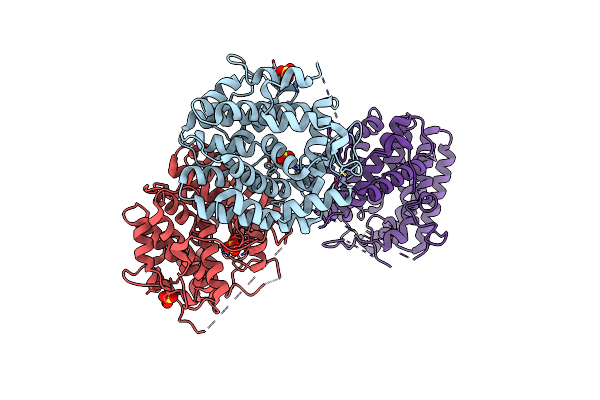 |
Organism: Shimazuella kribbensis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-12-11 Classification: LYASE Ligands: ZN, SO4 |
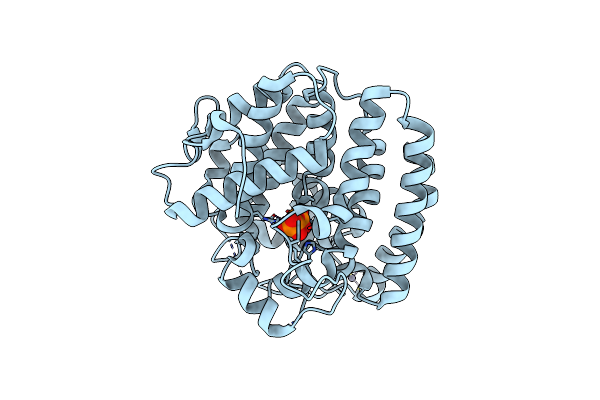 |
Crystal Structure Of Skaba3 From Shimazuella Kribbensis In Complex With Ppi
Organism: Shimazuella kribbensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-11 Classification: LYASE Ligands: ZN, MG, POP |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |