Search Count: 840
 |
Organism: Rubrivivax gelatinosus il144
Method: X-RAY DIFFRACTION Resolution:0.81 Å Release Date: 2018-02-07 Classification: UNKNOWN FUNCTION |
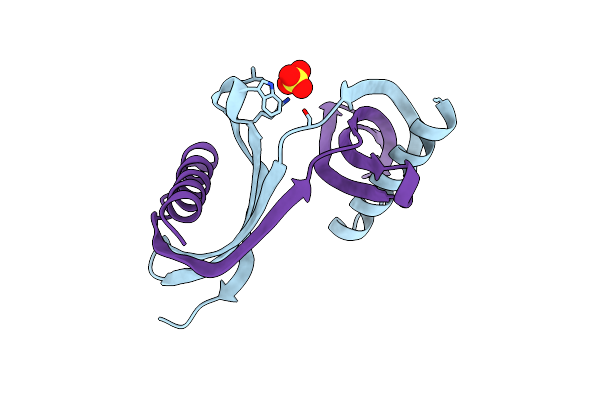 |
Crystal Structure Of A Single-Domain Cysteine Protease Inhibitor From Cowpea (Vigna Unguiculata)
Organism: Vigna unguiculata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-10-14 Classification: HYDROLASE INHIBITOR Ligands: SO4 |
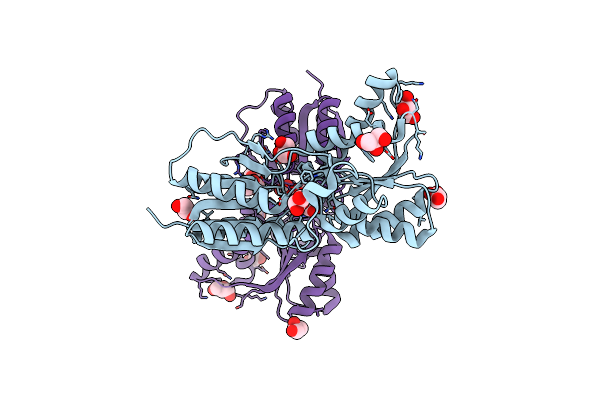 |
Crystal Structure Of Carbohydrate Transporter Solute Binding Protein Veis_2079 From Verminephrobacter Eiseniae Ef01-2, Target Efi-511009, A Complex With D-Talitol
Organism: Verminephrobacter eiseniae ef01-2
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-24 Classification: TRANSPORT PROTEIN Ligands: TLZ, GOL, BCT |
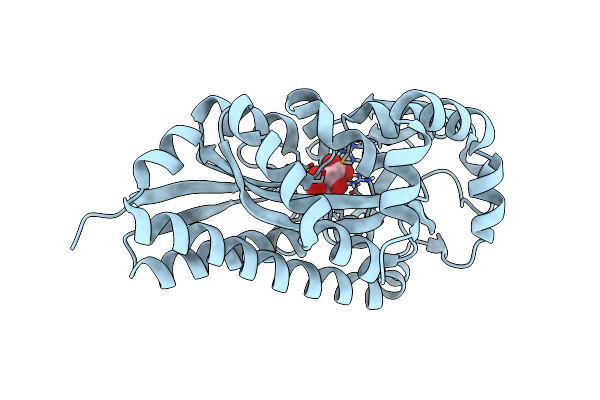 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Verminephrobacter Eiseniae Ef01-2 (Veis_3954, Target Efi-510324) A Nephridial Symbiont Of The Earthworm Eisenia Foetida, Bound To D-Erythronate With Residual Density Suggestive Of Superposition With Copurified Alternative Ligand.
Organism: Verminephrobacter eiseniae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-05-07 Classification: TRANSPORT PROTEIN Ligands: EAX |
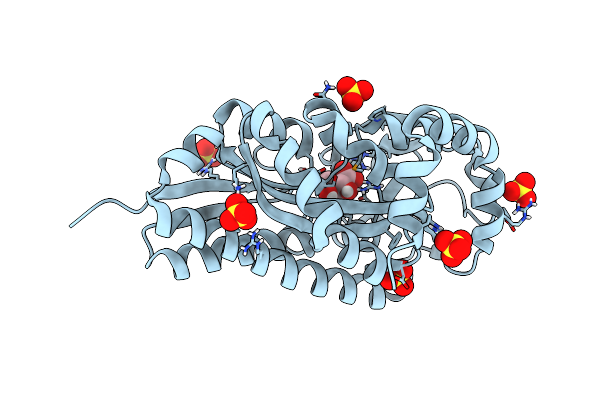 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Verminephrobacter Eiseniae Ef01-2 (Veis_3954, Target Efi-510324) A Nephridial Symbiont Of The Earthworm Eisenia Foetida, Bound To (R)-Pantoic Acid
Organism: Verminephrobacter eiseniae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-05-07 Classification: SOLUTE-BINDING PROTEIN Ligands: SO4, PAF |
 |
Crystal Structure Of Extracellular Ligand-Binding Receptor From Verminephrobacter Eiseniae Ef01-2
Organism: Verminephrobacter eiseniae
Method: X-RAY DIFFRACTION Release Date: 2013-11-06 Classification: TRANSPORT PROTEIN Ligands: GOL |
 |
Structure Of The Type Iva Major Pilin From The Electrically Conductive Bacterial Nanowires Of Geobacter Sulfurreducens
Organism: Geobacter sulfurreducens
Method: SOLUTION NMR Release Date: 2013-08-28 Classification: Cell Adhesion, Structural Protein, Electron Transport |
 |
Solution Structure Of A Mutant Of The Triheme Cytochrome Ppca From Geobacter Sulfurreducens Sheds Light On The Role Of The Conserved Aromatic Residue F15
Organism: Geobacter sulfurreducens
Method: SOLUTION NMR Release Date: 2013-01-30 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Crystal Structure Of A Fad-Dependent Pyridine Nucleotide-Disulphide Oxidoreductase From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: CA |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

