Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Comamonas testosteroni cnb-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-29 Classification: ISOMERASE |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-11-18 Classification: LIGASE |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-11-18 Classification: LIGASE Ligands: ACP |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-18 Classification: LIGASE Ligands: MG, ANP, MSL |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-11-18 Classification: LIGASE Ligands: MG, ADP, P3S |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-18 Classification: LIGASE Ligands: ADP |
 |
Organism: Rhodovulum sp. 12e13
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-18 Classification: LIGASE Ligands: ADP |
 |
Epsilon-Caprolactone-Bound Crystal Structure Of Cyclohexanone Monooxygenase In The Tight Conformation
Organism: Rhodococcus sp. hi-31
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2014-10-15 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, ECE, BCN |
 |
Epsilon-Caprolactone-Bound Crystal Structure Of Cyclohexanone Monooxygenase In The Loose Conformation
Organism: Rhodococcus sp. hi-31
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-10-15 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, ECE, PTD |
 |
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-07-02 Classification: HYDROLASE INHIBITOR Ligands: NI |
 |
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-07-02 Classification: HYDROLASE |
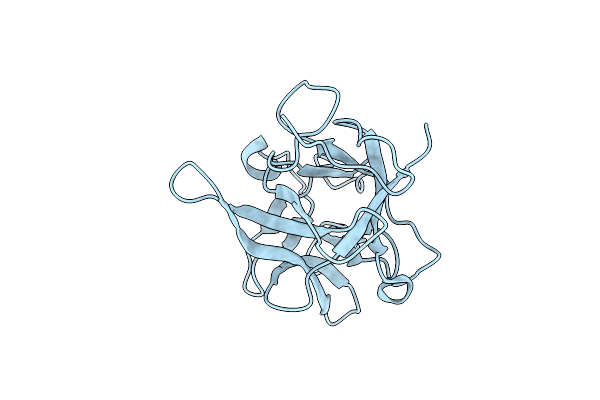 |
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-04-09 Classification: HYDROLASE INHIBITOR |
 |
A Binary Complex Betwwen Bovine Pancreatic Trypsin And A Engineered Mutant Trypsin Inhibitor
Organism: Bos taurus, Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2012-10-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Cyclohexanone-Bound Crystal Structure Of Cyclohexanone Monooxygenase In The Rotated Conformation
Organism: Rhodococcus sp. hi-31
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2012-04-25 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, CYH |
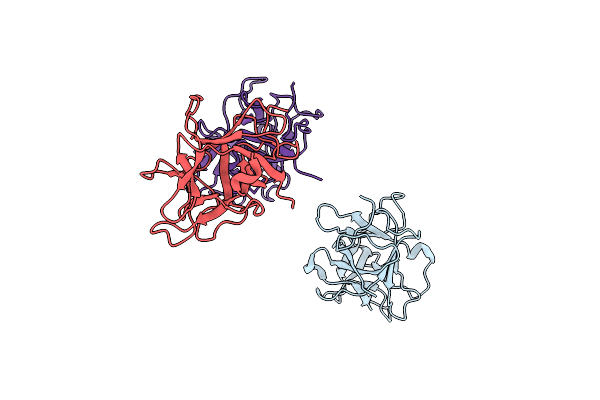 |
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2011-05-25 Classification: HYDROLASE INHIBITOR |
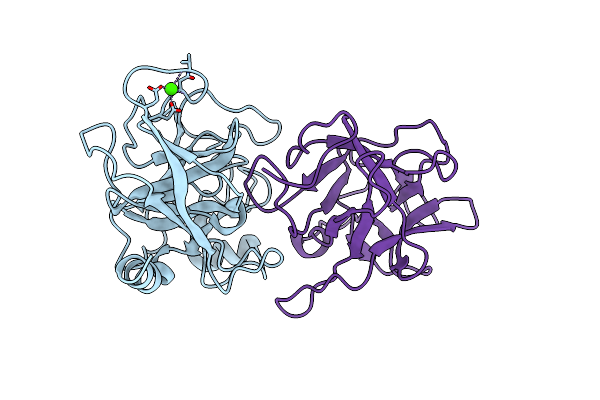 |
Crystal Structure Of A Binary Complex Between An Mutant Trypsin Inhibitor With Bovine Trypsin
Organism: Bos taurus, Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-06-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
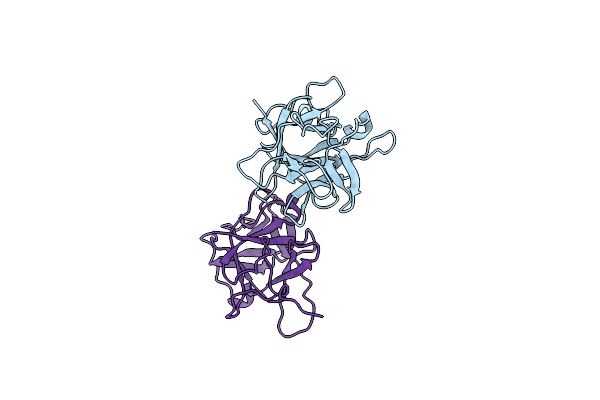 |
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-02 Classification: HYDROLASE INHIBITOR Ligands: CL |
 |
Crystal Structure Of A Chimeric Trypsin Inhibitor Having Reactive Site Loop Of Eti On The Scaffold Of Wci
Organism: Psophocarpus tetragonolobus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2010-02-02 Classification: HYDROLASE INHIBITOR |
 |
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-05-05 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |
 |
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-05 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |