Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
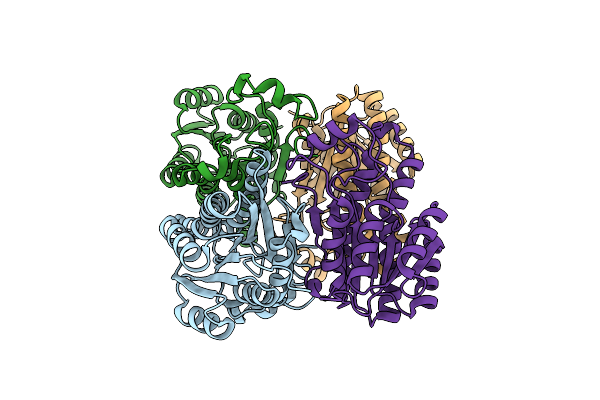
Organism: Levilactobacillus brevis atcc 367
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: HYDROLASE/TRANSPORT PROTEIN |
 |
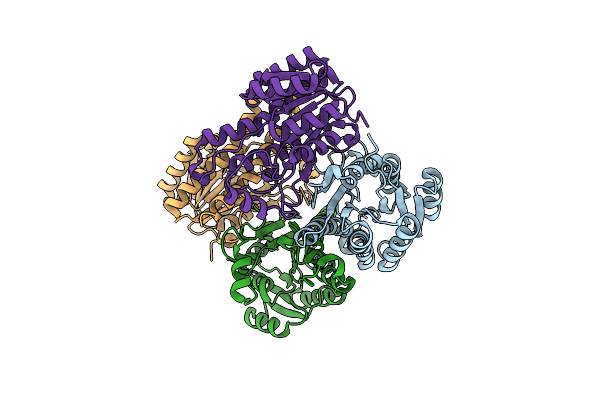
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE |
 |
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE |
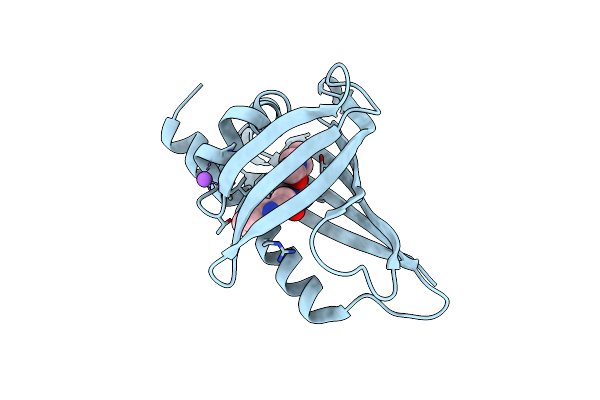 |
Crystal Structure Of Yellow Lupin Llpr-10.2B Protein In Complex With Melatonin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2018-04-18 Classification: PLANT PROTEIN Ligands: UNL, ML1, NA |
 |
Crystal Structure Of Yellow Lupin Llpr-10.2B Protein In Complex With Melatonin And Trans-Zeatin.
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2018-04-18 Classification: PLANT PROTEIN Ligands: UNL, NA, ML1, ZEA |
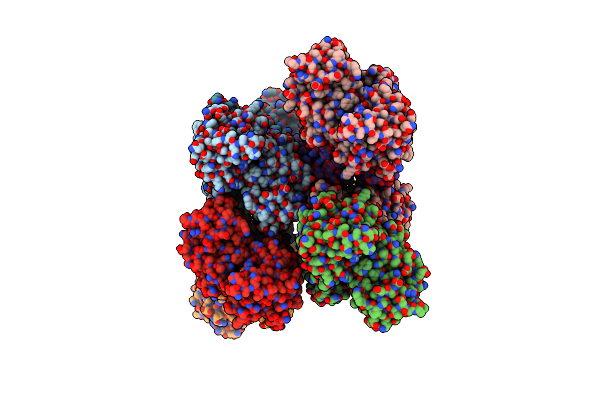 |
Water-Forming Nadh Oxidase From Lactobacillus Brevis (Lbnox) Bound To Nadh.
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-02 Classification: OXIDOREDUCTASE Ligands: FAD, OXY, NAI, EDO |
 |
Crystal Structure Of Dah7Ps-Cm Complex From Geobacillus Sp. With Prephenate
Organism: Geobacillus sp. ghh01
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-09-07 Classification: TRANSFERASE,ISOMERASE Ligands: MN, SO4, PRE |
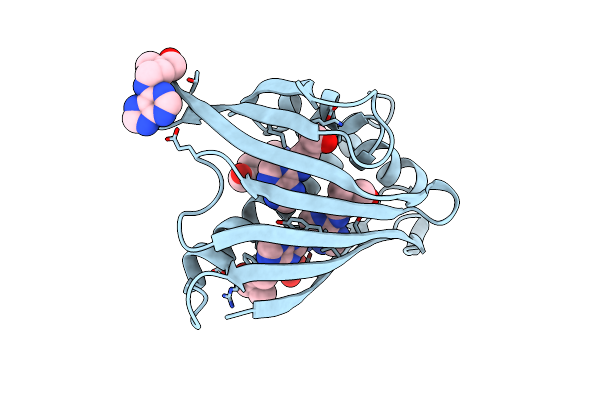 |
Crystal Structure Of Yellow Lupin Llpr-10.1A Protein In Complex With Trans-Zeatin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN Ligands: ZEA, SO4 |
 |
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN Ligands: ACT |
 |
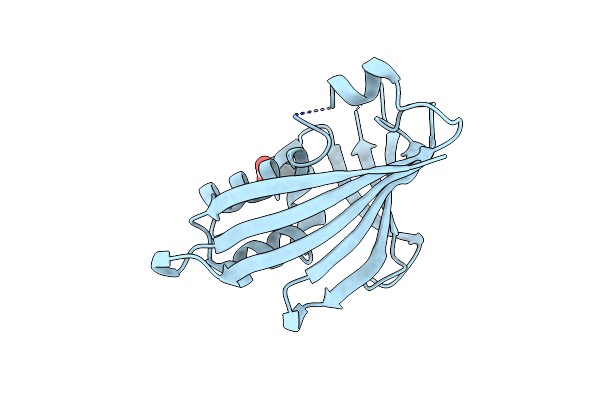
Crystal Structure Of Yellow Lupine Llpr-10.1A Protein Partially Saturated With Trans-Zeatin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN |
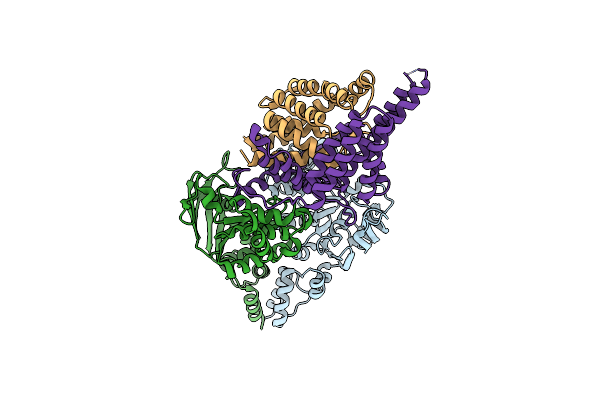 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-04-17 Classification: HYDROLASE |
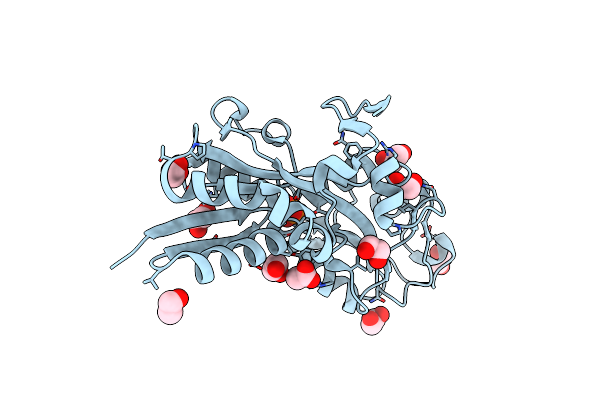 |
Crystal Structure Of An Abc-Type Phosphate Transport System, Periplasmic Component (Lvis_0633) From Lactobacillus Brevis Atcc 367 At 1.55 A Resolution
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-04-11 Classification: PHOSPHATE-BINDING PROTEIN Ligands: PO4, ACT, EDO |
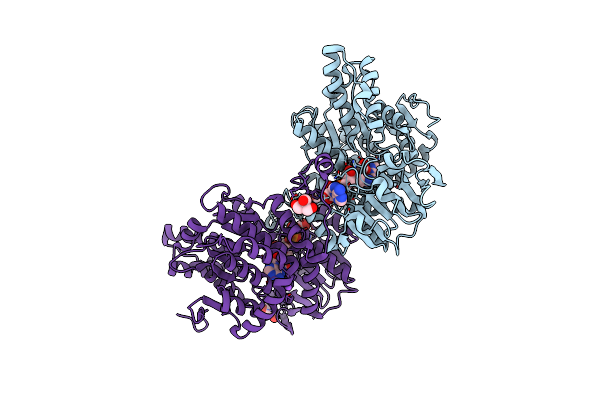 |
Crystal Structure Of Lupinus Luteus S-Adenosyl-L-Homocysteine Hydrolase In Complex With Adenosine
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2011-08-31 Classification: PLANT PROTEIN Ligands: NAD, TRS, ADN, NA |
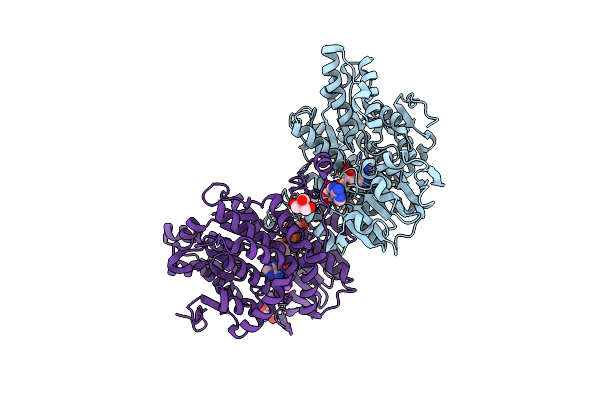 |
Crystal Structure Of Lupinus Luteus S-Adenosyl-L-Homocysteine Hydrolase In Complex With Adenine
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2011-08-31 Classification: HYDROLASE/HYDROLASE SUBSTRATE Ligands: NAD, TRS, ADE, NA |
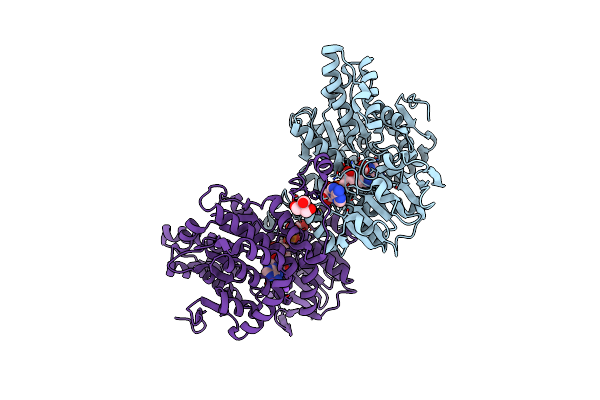 |
Crystal Structure Of Lupinus Luteus S-Adenosyl-L-Homocysteine Hydrolase In Complex With Cordycepin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAD, 3AD, NA, TRS |
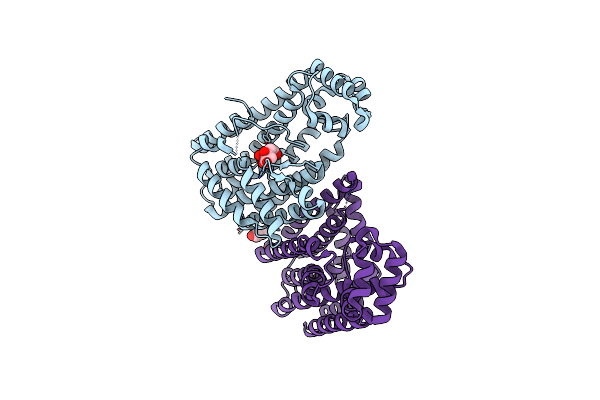 |
Crystal Structure Of Geranylgeranyl Pyrophosphate Synthase From Lactobacillus Brevis Atcc 367 Complexed With Citrate
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2010-11-24 Classification: TRANSFERASE Ligands: CIT, GOL |
 |
Crystal Structure Of Geranylgeranyl Pyrophosphate Synthase From Lactobacillus Brevis Atcc 367 Complexed With Isoprenyl Diphosphate And Magnesium
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-11-17 Classification: TRANSFERASE Ligands: DMA, MG, GOL |
 |
Crystal Structure Of Geranylgeranyl Pyrophosphate Synthase From Lactobacillus Brevis Atcc 367
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-06-02 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-05-12 Classification: HYDROLASE |
 |
Crystal Structure Of The Alpha-Galactosidase From Lactobacillus Brevis, Northeast Structural Genomics Consortium Target Lbr11.
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-04-28 Classification: HYDROLASE |