Search Count: 365
 |
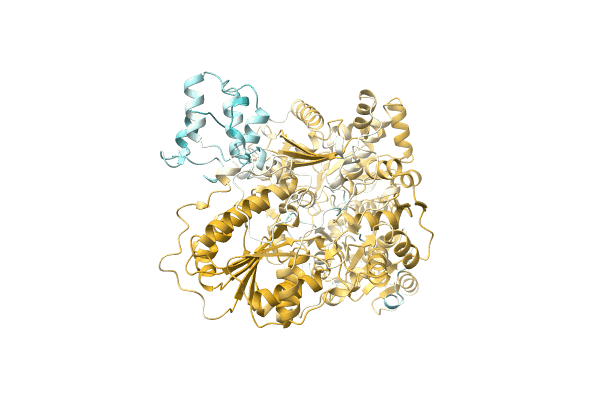
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Prevotella Timonensis
Organism: Prevotella timonensis
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: EPE, SO4 |
 |
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Prevotella Timonensis
Organism: Prevotella timonensis
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, EDO |
 |
Organism: Lens culinaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-03-09 Classification: ALLERGEN |
 |
Spatial Structure Of The Lentil Lipid Transfer Protein In Complex With Anionic Lysolipid Lppg
Organism: Lens culinaris
Method: SOLUTION NMR Release Date: 2017-06-28 Classification: LIPID TRANSPORT Ligands: PGM |
 |
Organism: Lens culinaris
Method: SOLUTION NMR Release Date: 2013-10-02 Classification: LIPID TRANSPORT |
 |
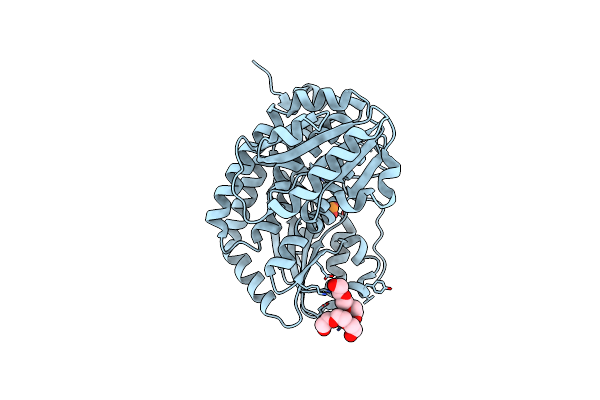
Crystal Structure Of A Putative Conserved Lipoprotein (Nt01Cx_1156) From Clostridium Novyi Nt At 2.70 A Resolution
Organism: Clostridium novyi
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-06-26 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
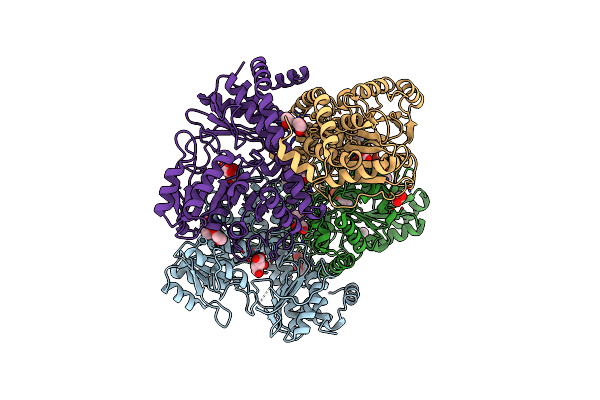
Crystal Structure Of A Putative Tagatose-6-Phosphate Ketose/Aldose Isomerase (Nt01Cx_0292) From Clostridium Novyi Nt At 2.35 A Resolution
Organism: Clostridium novyi
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-12-01 Classification: ISOMERASE Ligands: PG4, PO4 |
 |
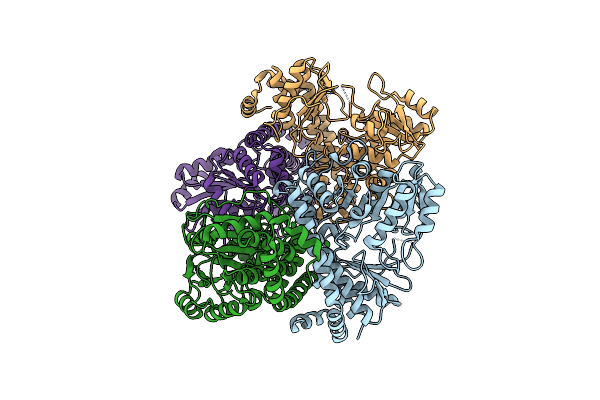
Crystal Structure Of Carbon-Sulfur Lyase Involved In Aluminum Resistance (Yp_878183.1) From Clostridium Novyi Nt At 2.00 A Resolution
Organism: Clostridium novyi
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-07-21 Classification: LYASE Ligands: TLA, EDO, PLP |
 |
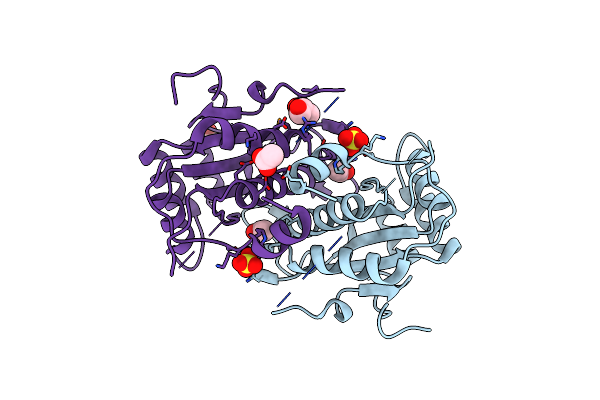
Crystal Structure Of Carbon-Sulfur Lyase Involved In Aluminum Resistance (Yp_878183.1) From Clostridium Novyi Nt At 2.90 A Resolution
Organism: Clostridium novyi nt
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-04-14 Classification: LYASE |
 |
Crystal Structure Of Nitroreductase Family Protein (Yp_877874.1) From Clostridium Novyi Nt At 1.75 A Resolution
Organism: Clostridium novyi
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-02-17 Classification: OXIDOREDUCTASE Ligands: SO4, ACT, GOL |
 |
The Monosaccharide Binding Site Of Lentil Lectin: An X-Ray And Molecular Modelling Study
Organism: Lens culinaris
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-01-31 Classification: LECTIN Ligands: GLC, CA, MN |
 |
Refinement Of Two Crystal Forms Of Lentil Lectin At 1.8 Angstroms Resolution
Organism: Lens culinaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: LECTIN Ligands: PO4, MN, CA |
 |
Crystal Structure Determination And Refinement At 2.3 Angstroms Resolution Of The Lentil Lectin
Organism: Lens culinaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: LECTIN Ligands: PO4, MN, CA |
 |
 |
 |
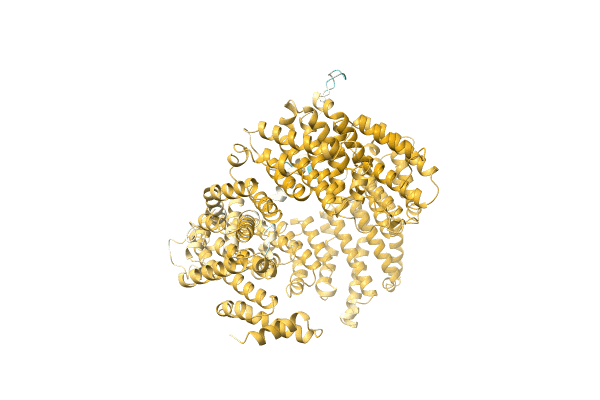 |
 |
 |
 |

