Search Count: 2,777
 |
Co-Crystal Structure Of Feline Coronavirus Uu23 Main Protease With Pfizer Intravenous Compound Pf-00835231
Organism: Feline alphacoronavirus 1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: V2M, ACY, PG4, EDO, PEG |
 |
N-Acyl-D-Amino-Acid Deacylase (D-Acylase) From Klebsiella Pneumoniae In An Open Conformation
Organism: Klebsiella pneumoniae subsp. pneumoniae kp13
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: SO4, PEG, EDO, GOL, PGE, NI |
 |
N-Acyl-D-Amino-Acid Deacylase (D-Acylase) From Klebsiella Pneumoniae In The Absence Of Glycerol
Organism: Klebsiella pneumoniae subsp. pneumoniae kp13
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: EDO, SO4, NI |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Organism: Bacillus, Bacillus phage spo1
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: TRANSCRIPTION |
 |
Organism: Bacillus, Bacillus phage spo1
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: TRANSCRIPTION |
 |
Organism: Bacillus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
His579Leu Variant Of L-Arabinonate Dehydratase Co-Crystallized With 2-Oxobutyrate
Organism: Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-09-25 Classification: LYASE Ligands: MG, FES, 2KT |
 |
Organism: Streptomyces yokosukanensis
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-06-05 Classification: BIOSYNTHETIC PROTEIN Ligands: YG3, DMS, SO4, GOL |
 |
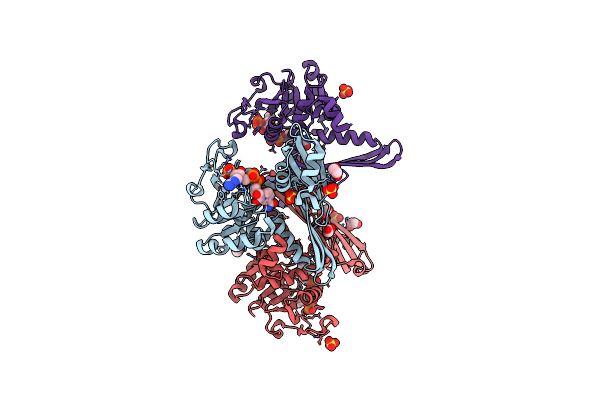
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Prevotella Timonensis
Organism: Prevotella timonensis
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: EPE, SO4 |
 |
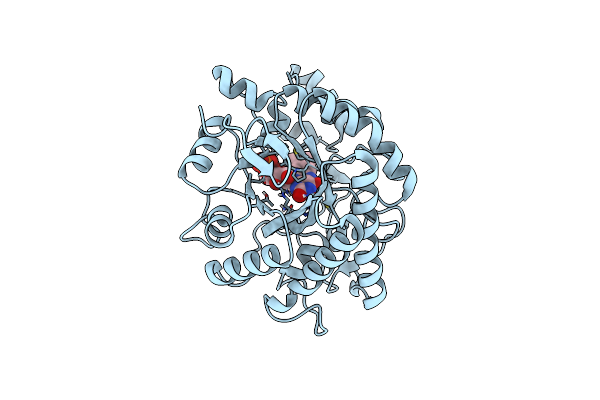
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Prevotella Timonensis
Organism: Prevotella timonensis
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, EDO |
 |
Organism: Stutzerimonas chloritidismutans aw-1
Method: X-RAY DIFFRACTION Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: FNR, CL |
 |
Organism: Escherichia coli o157:h7 str. sakai, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: AYE |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In A Nanodisc
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: TRANSPORT PROTEIN Ligands: OCT, PTY, TWT, D10, NA, TRD, HEX |
 |
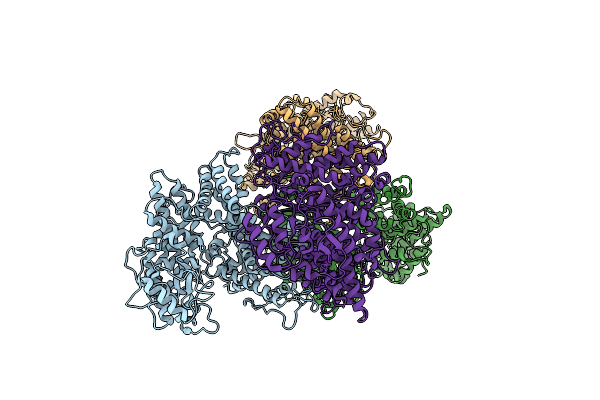
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In Amphipol
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
 |
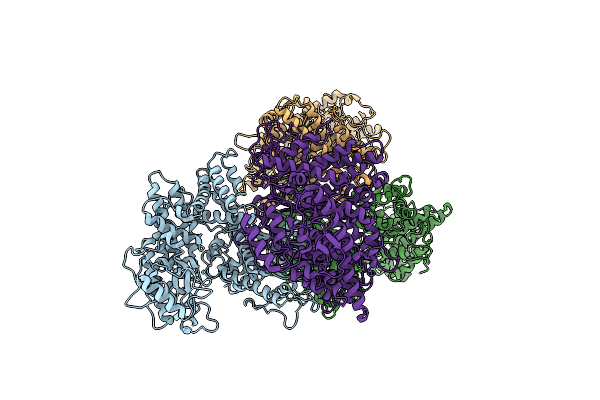
Organism: Streptomyces yokosukanensis
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2022-06-29 Classification: HYDROLASE |
 |
Organism: Streptomyces yokosukanensis
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2022-06-29 Classification: HYDROLASE |
 |
Organism: Lens culinaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-03-09 Classification: ALLERGEN |

