Search Count: 2,474
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: CA |
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: ADP |
 |
Nuclear Export Protein/Non-Structural Protein 2 (Nep/Ns2) In Complex With Artificial Alpha Rep Protein
Organism: Synthetic construct, Influenza a virus (a/wsn/1933(h1n1))
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Aeromonas Salmonicida Putative Carbohydrate Binding Module And Split Domain
Organism: Aeromonas salmonicida subsp. salmonicida a449
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: UDP, A1EGV |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: HW2, UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: J6O, UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: U5P, RUT |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGW, UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGX, UDP |
 |
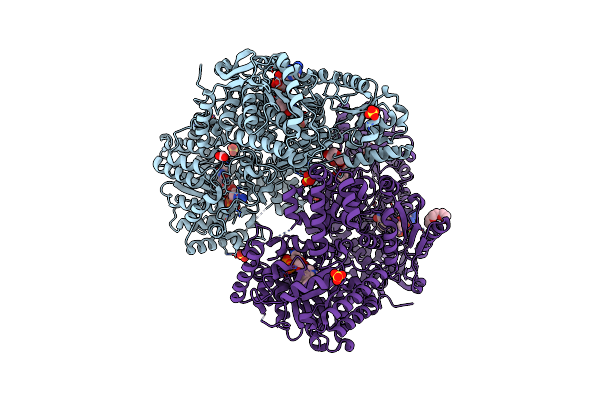
Proline Utilization A Complexed With The Product L-Glutamate In The Aldehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: GGL, SO4, PGE, FAD, PEG |
 |
Proline Utilization A (Puta) From Sinorhizobium Meliloti Inactivated By N-Propargylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: P5F, NAD, SO4, MG, FMT, PEG, PG4, PGE |
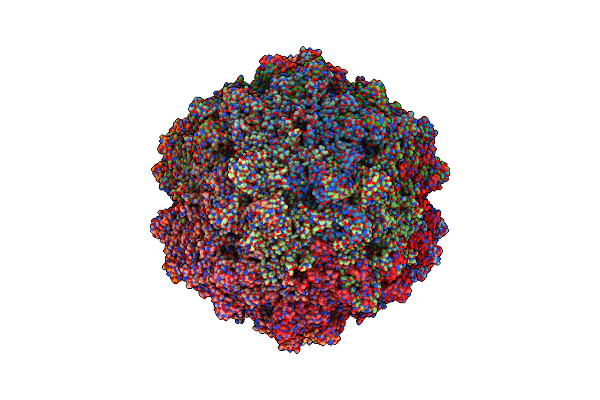 |
Organism: Goose parvovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRUS LIKE PARTICLE |
 |
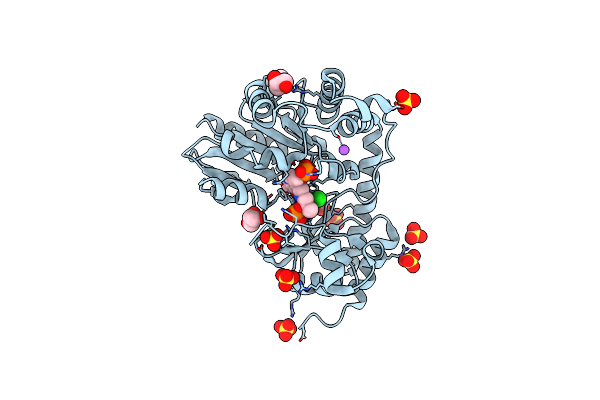
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-16 Classification: LYASE Ligands: GOL, SO4, CL, NA |
 |
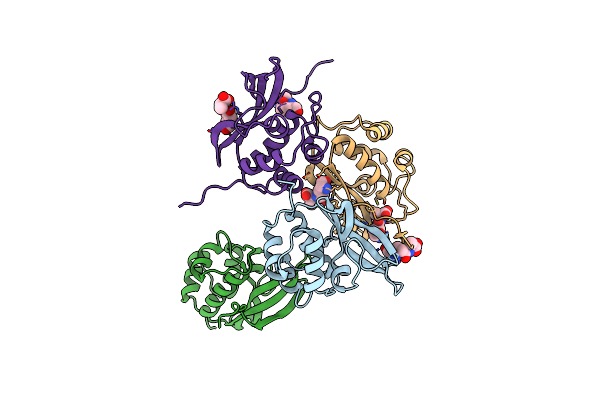
Crystal Structure Of L-Threonine-O-3-Phosphate Decarboxylase Cobc In Complex With Reaction Intermediate
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-10-16 Classification: LYASE Ligands: GOL, 33P, SO4, CL, NA |
 |
Organism: Methanothermobacter phage psim100
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: DGL, ALA |
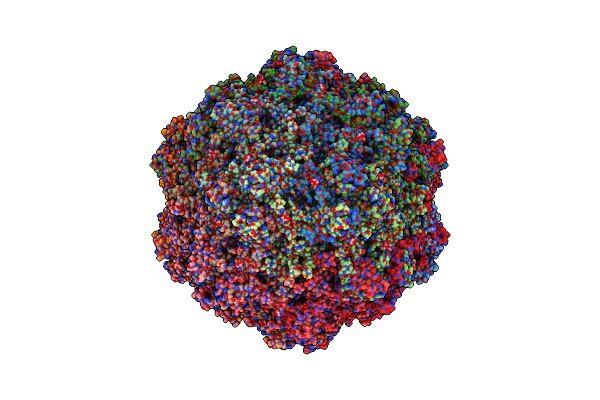 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRUS LIKE PARTICLE Ligands: SIA |
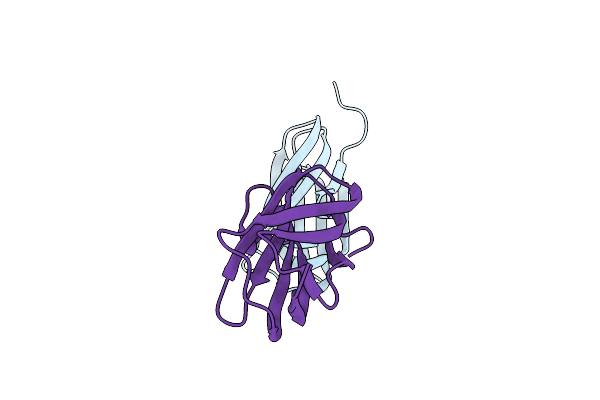 |
Organism: Flammeovirga sp. oc4
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-11 Classification: SUGAR BINDING PROTEIN |
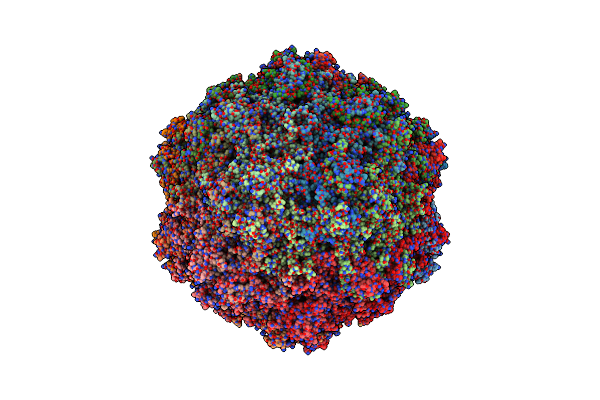 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |

