Search Count: 518
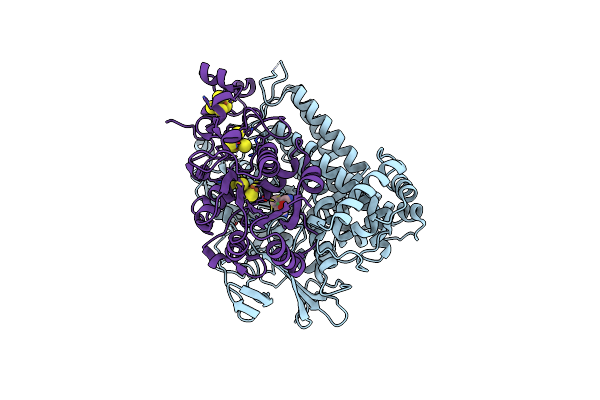 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.93 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, 35L, SF3, CL |
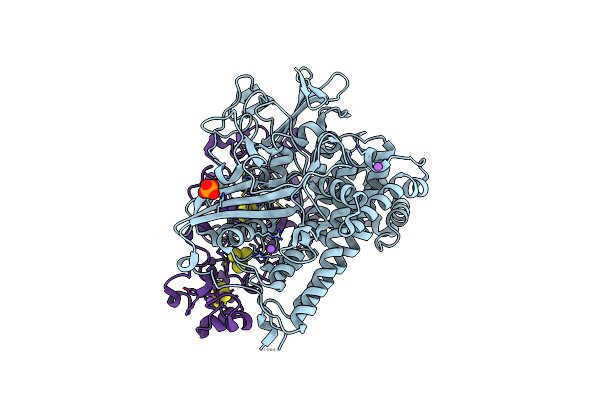 |
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.6 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, NA, PO4, SF4, F3S, CL, ER2 |
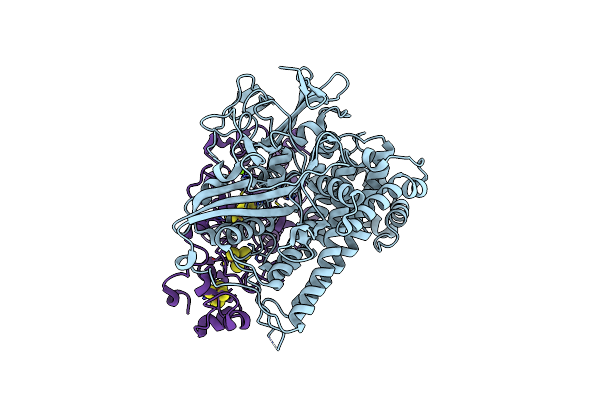 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, CL, SF4, F3S, 35L, F4S |
 |
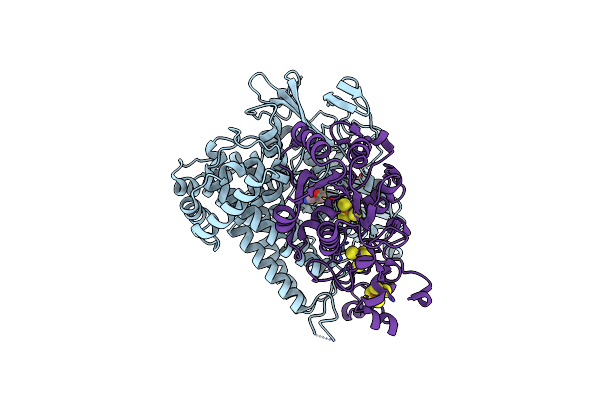
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.61 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S, ER2 |
 |
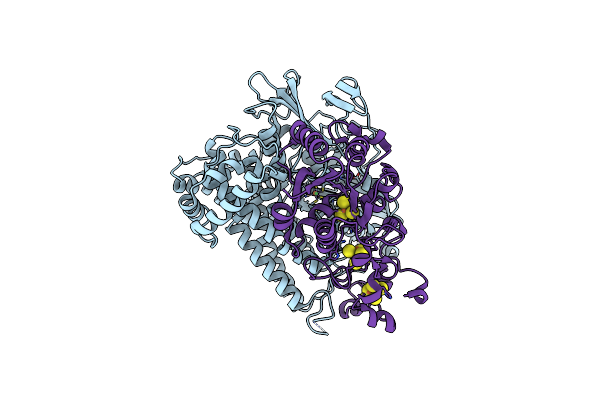
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S |
 |
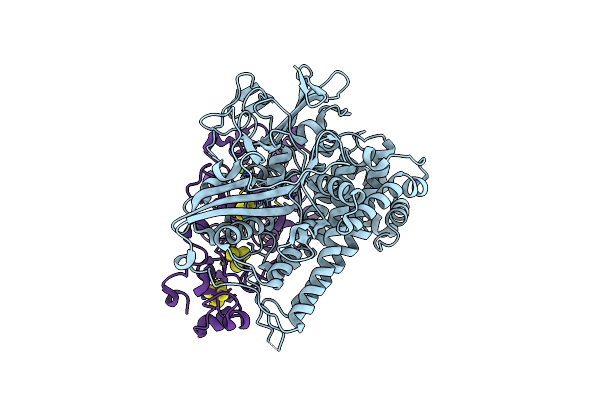
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.92 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFU, MG, SF4, F3S |
 |
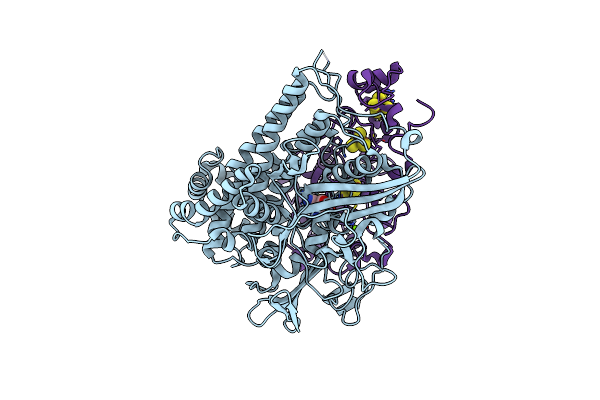
Crystal Structure Of The O2-Tolerant Mbh-P242C From Ralstonia Eutropha In Its Reduced State
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337), Cupriavidus necator (strain atcc 17699 / dsm 428 / kctc 22496 / ncimb 10442 / h16 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-07-07 Classification: ELECTRON TRANSPORT Ligands: NFU, MG, SF4, F4S, CL |
 |
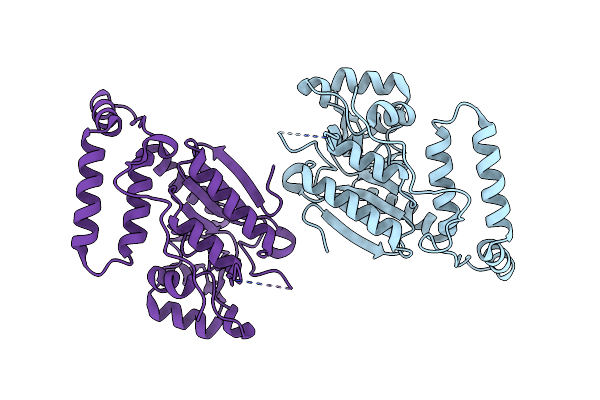
Crystal Structure Of The O2-Tolerant Mbh-P242C From Ralstonia Eutropha In Its As-Isolated State
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337), Cupriavidus necator (strain atcc 17699 / dsm 428 / kctc 22496 / ncimb 10442 / h16 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2021-07-07 Classification: ELECTRON TRANSPORT Ligands: NFV, CL, MG, SF4, O, F4S |
 |
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-08-21 Classification: LYASE |
 |
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: GOL, PO4 |
 |
Crystal Structure Of Enoyl-Coa Hydratase (Ech) From Ralstonia Eutropha H16 In Complex With Nad
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: GOL, NAD |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In A Its As-Isolated High-Pressurized Form
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, F4S, CL |
 |
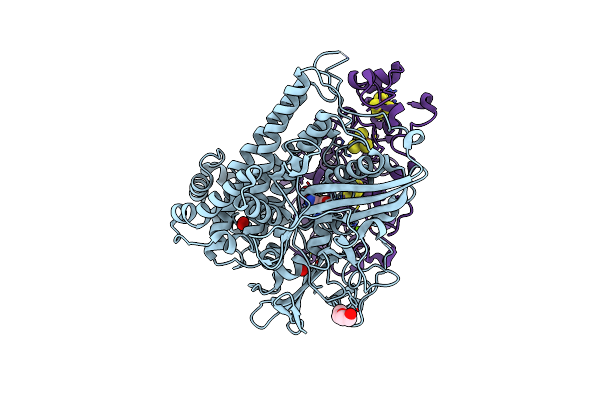
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form (Oxidized State - State 3)
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: MG, CL, NFV, SF4, F3S, F4S |
 |
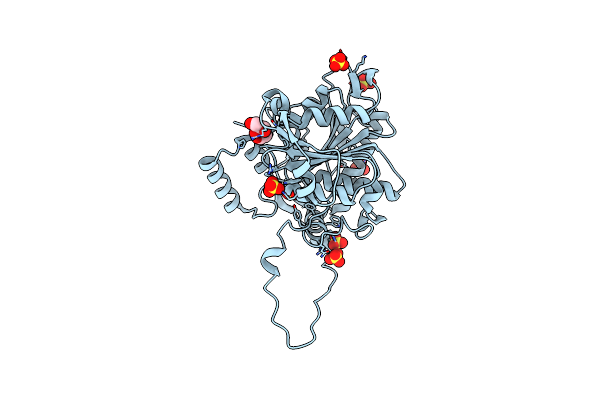
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its O2-Derivatized Form By A "Soak-And-Freeze" Derivatization Method
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, OXY, PEG, SF4, F3S, F4S, CL, O |
 |
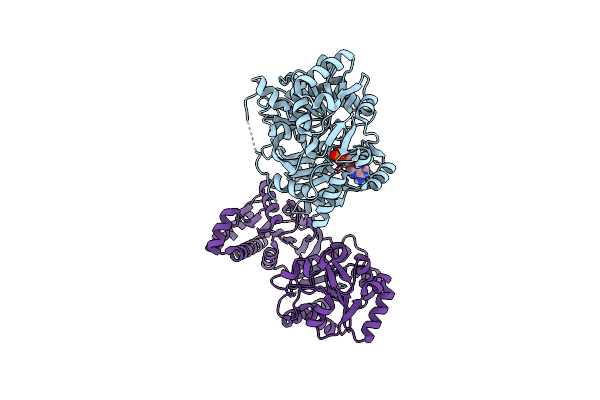
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-12-07 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
Crystal Structure Of A Domain Of Unknown Function (Duf1537) From Ralstonia Eutropha H16 (H16_A1561), Target Efi-511666, Complex With Adp.
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-10-28 Classification: UNKNOWN FUNCTION Ligands: ADP |
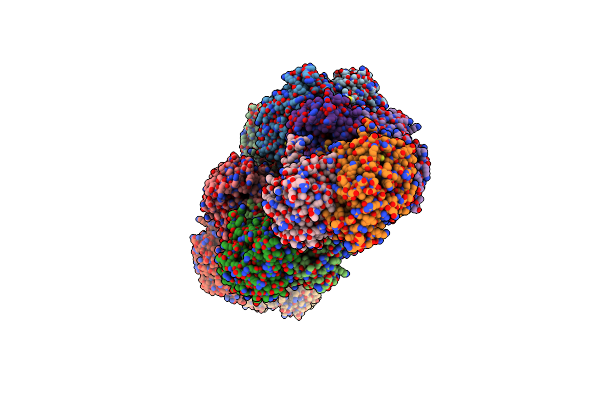 |
Crystal Structure Of Beta-Ketoacyl Thiolase B (Bktb) From Ralstonia Eutropha
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-09-09 Classification: TRANSFERASE |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 In Complex With Acetoacetyl-Coa
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-04-22 Classification: OXIDOREDUCTASE Ligands: CAA |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 From Ralstonia Eutropha
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-02-11 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 In Complex With Nad+
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2015-02-11 Classification: OXIDOREDUCTASE Ligands: NAD |

