Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
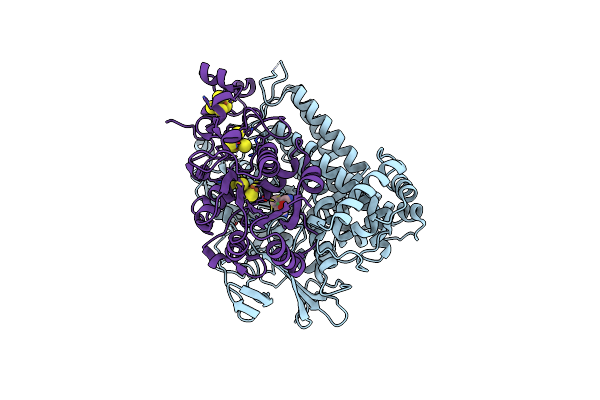 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.93 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, 35L, SF3, CL |
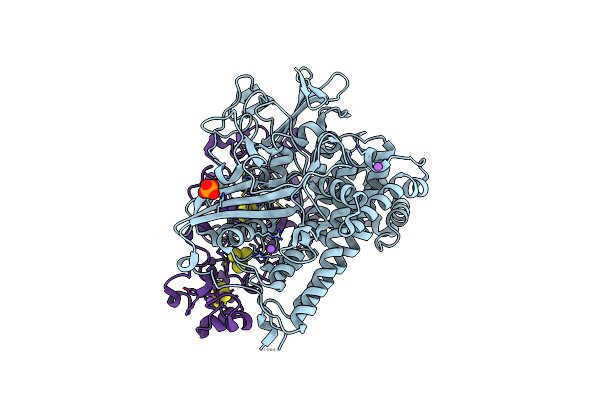 |
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.6 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, NA, PO4, SF4, F3S, CL, ER2 |
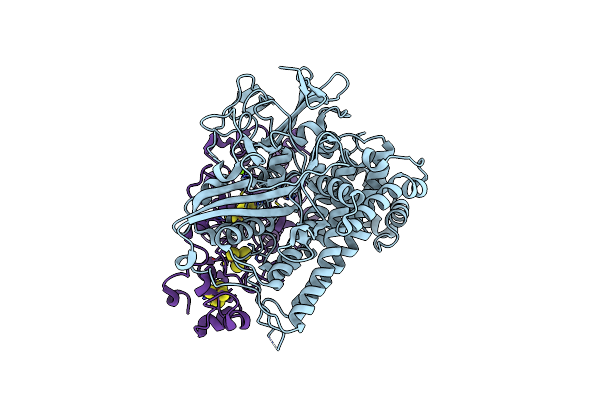 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, CL, SF4, F3S, 35L, F4S |
 |
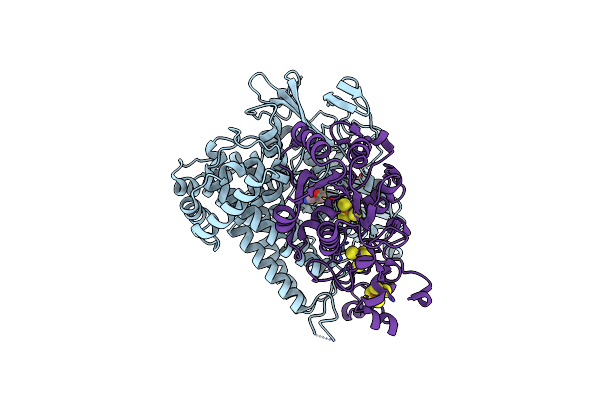
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.61 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S, ER2 |
 |
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S |
 |
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.92 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFU, MG, SF4, F3S |
 |
Organism: Thermoplasmatales archaeon sg8-52-1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-09-07 Classification: MEMBRANE PROTEIN Ligands: RET, CL, D12 |
 |
Organism: Abrus precatorius
Method: X-RAY DIFFRACTION Release Date: 2022-06-15 Classification: HYDROLASE Ligands: COA |
 |
Crystal Structure Of The O2-Tolerant Mbh-P242C From Ralstonia Eutropha In Its Reduced State
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337), Cupriavidus necator (strain atcc 17699 / dsm 428 / kctc 22496 / ncimb 10442 / h16 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-07-07 Classification: ELECTRON TRANSPORT Ligands: NFU, MG, SF4, F4S, CL |
 |
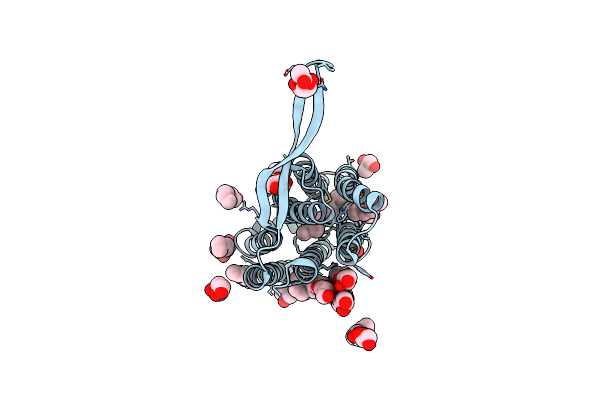
Crystal Structure Of The O2-Tolerant Mbh-P242C From Ralstonia Eutropha In Its As-Isolated State
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337), Cupriavidus necator (strain atcc 17699 / dsm 428 / kctc 22496 / ncimb 10442 / h16 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2021-07-07 Classification: ELECTRON TRANSPORT Ligands: NFV, CL, MG, SF4, O, F4S |
 |
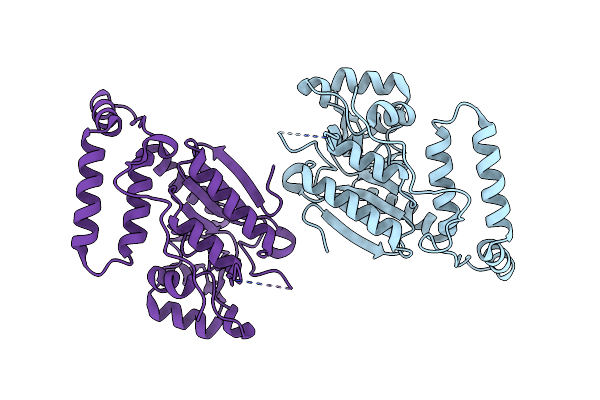
Crystal Structure Of Thermoplasmatales Archaeon Heliorhodopsin E108D Mutant
Organism: Thermoplasmatales archaeon sg8-52-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-09-02 Classification: MEMBRANE PROTEIN Ligands: RET, OLC, SO4 |
 |
Organism: Thermoplasmatales archaeon sg8-52-1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-09-25 Classification: MEMBRANE PROTEIN Ligands: OLC, RET |
 |
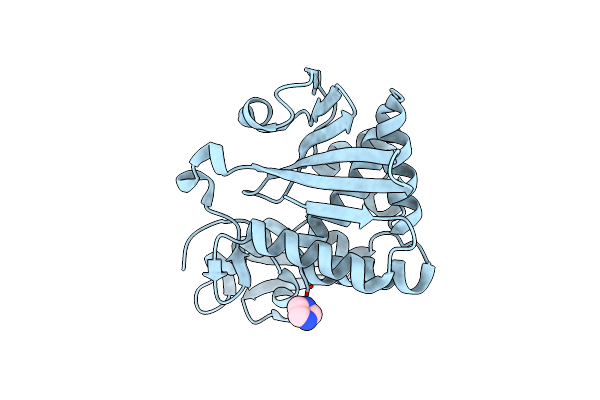
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-08-21 Classification: LYASE |
 |
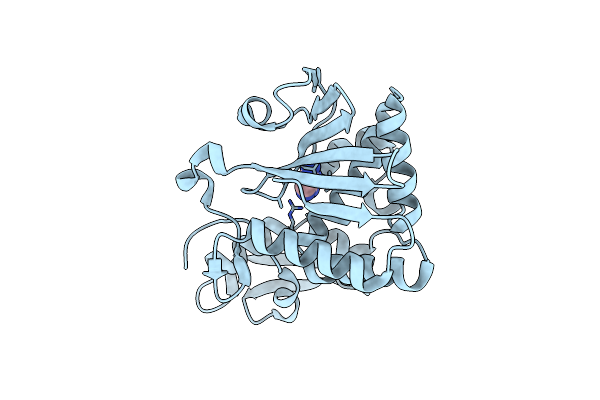
Organism: Abrus precatorius
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-01-02 Classification: HYDROLASE Ligands: IMD |
 |
Crystal Structure Of Abrin A Chain (Recombinant) In Complex With Adenine At 1.65 Angstroms
Organism: Abrus precatorius
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-01-02 Classification: HYDROLASE Ligands: ADE |
 |
Crystal Structure Of Abrin A Chain (Recombinant) In Complex With Nicotinamide At 1.7 Angstroms
Organism: Abrus precatorius
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-02 Classification: HYDROLASE Ligands: IMD, NCA |
 |
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: GOL, PO4 |
 |
Crystal Structure Of Enoyl-Coa Hydratase (Ech) From Ralstonia Eutropha H16 In Complex With Nad
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: GOL, NAD |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In A Its As-Isolated High-Pressurized Form
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, F4S, CL |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form (Oxidized State - State 3)
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: MG, CL, NFV, SF4, F3S, F4S |