Search Count: 3,086
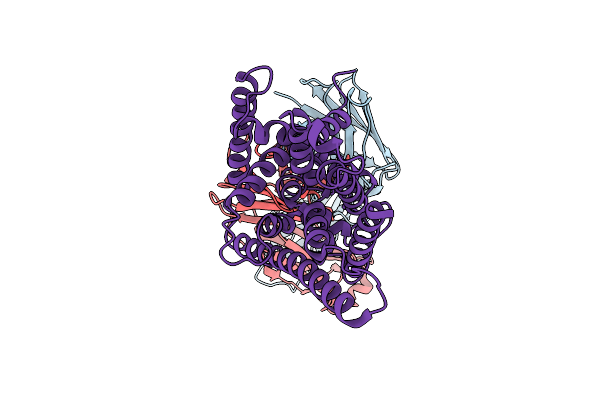 |
Cryo-Em Structure Of Candida Albicans Fluoride Channel Fex In Complex With Fab Fragment
Organism: Homo sapiens, Hiv-1 m:b_mn, Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
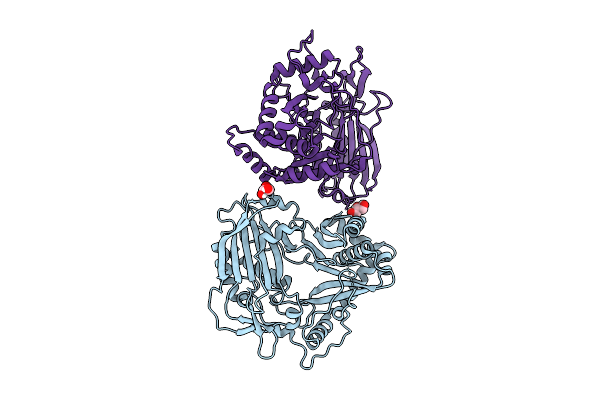 |
Organism: Echinacea purpurea
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GOL |
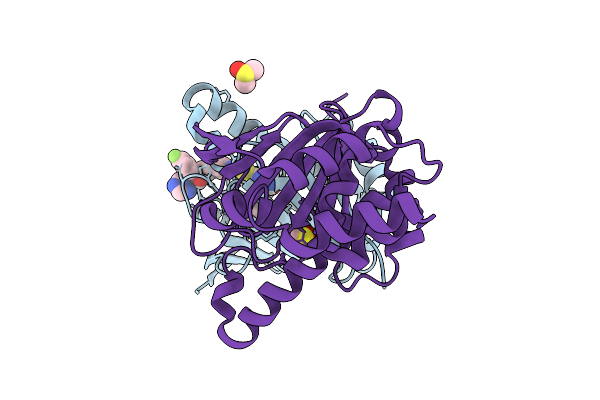 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: CHAPERONE Ligands: A1A27, DMS, CL |
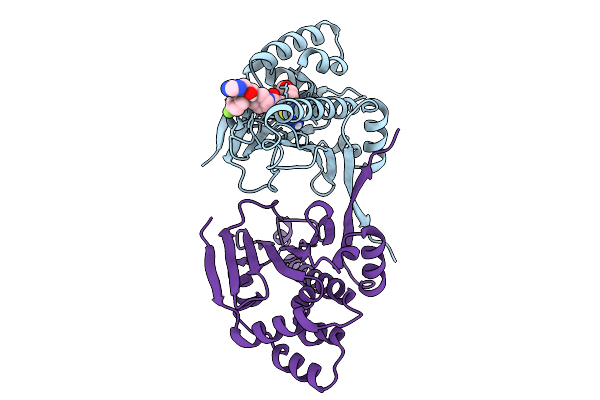 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A28 |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A26, MG |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A24, MG |
 |
A Vast Marine Sulfonate-Based Carbon Cycle Fueled By Novel Sulfoquinovosidases
Organism: Roseobacter denitrificans och 114
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PROTEIN BINDING Ligands: NAD, PEG |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With 4456
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYI |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Sj6675
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYV |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: EDO, ACT, MPD, PEG, BTB |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: EDO, GOL, MPD |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: MPD, EDO, BTB, PEG, ACT |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: MPD, BTB, EDO |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: PEG, MPD, BTB, GOL, EDO |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: EDO, MPD, PEG |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: EDO, MPD, PEG |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: MPD, EDO, ACT, GOL |
 |
Organism: Lucilia cuprina
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: MPD, EDO |
 |
Crystal Structure Of Biphenyl Synthase From Malus Domestica Complexed With Diketide-Coa Mimetics
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BU4, A1AT9, A1AT8 |
 |
Crystal Structure Of Biphenyl Synthase From Malus Domestica Complexed With Triketide-Coa Mimetic
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BU4, A1AUA |

