Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
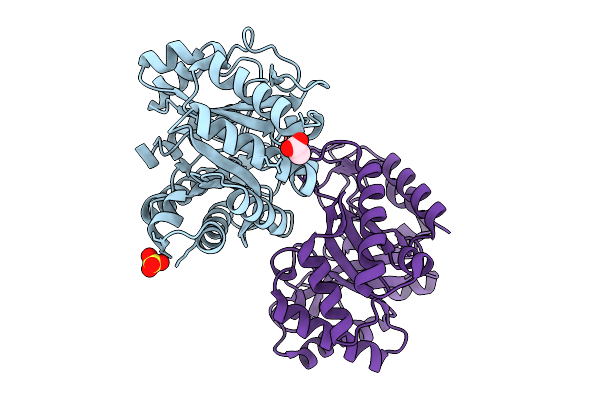
Crystal Structure Of Egtuc Binding Domain Double Mutant I243P T274G Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN Ligands: LW8, FLC, CL, NA |
 |
Organism: Feline infectious peritonitis virus (strain 79-1146), Felis catus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: NAG |
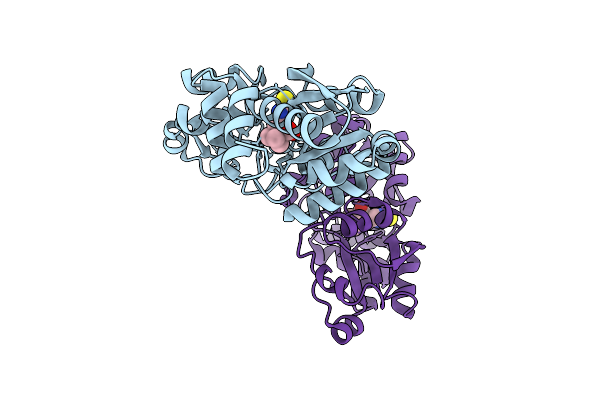 |
Crystal Structure Of Egtuc Binding Domain Mutant I243A Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8 |
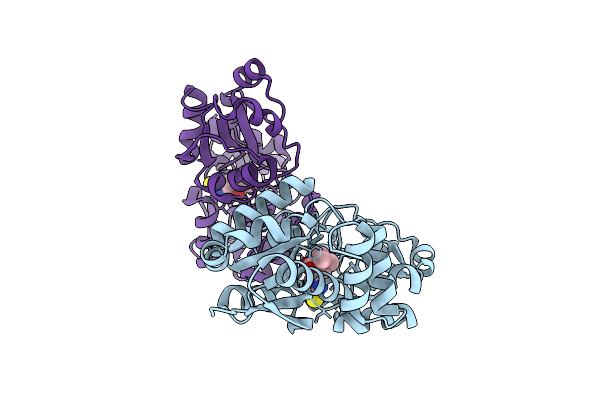 |
Crystal Structure Of Egtuc Binding Domain Mutant T274G Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8 |
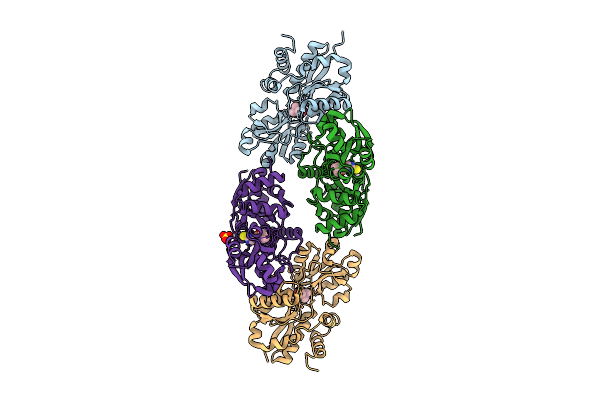 |
Crystal Structure Of Egtuc Binding Domain Mutant I243P Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8, SO4 |
 |
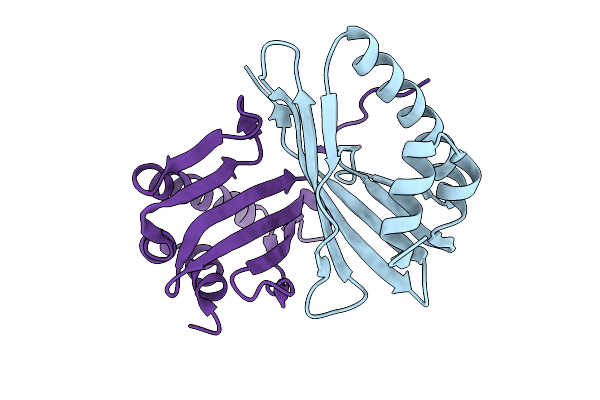
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: SO4, ACT |
 |
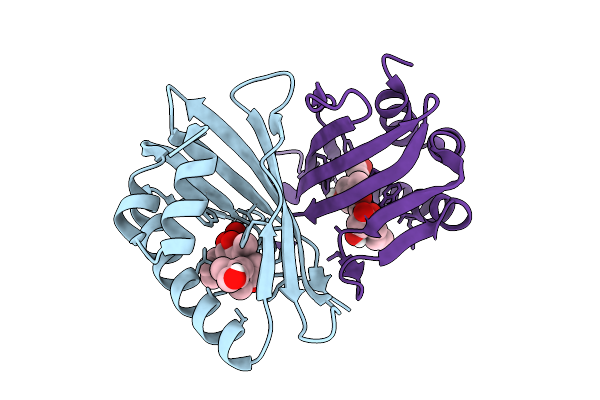
Organism: Saccharothrix syringae
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
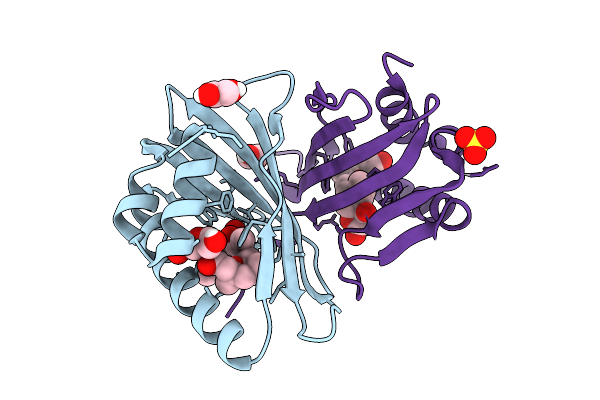
Organism: Saccharothrix syringae
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: A1D9Z |
 |
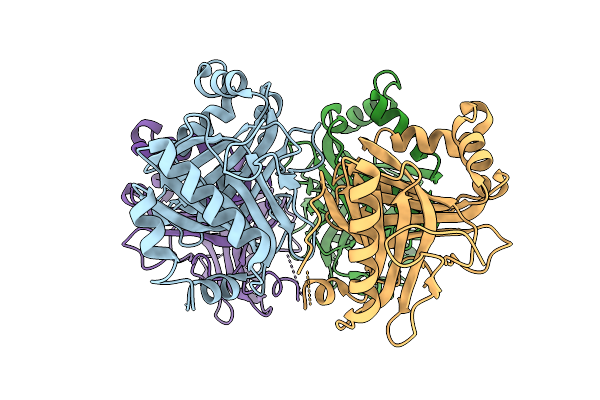
Organism: Saccharothrix syringae
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: A1D90, EDO, PEG, SO4 |
 |
Organism: Saccharothrix espanaensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE |
 |
Organism: Hordeum vulgare, Blumeria graminis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: ANTIFUNGAL PROTEIN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Target-Cleaved State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Target-Released State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, DC |
 |
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: DNA BINDING PROTEIN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Rna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Pre-Cleavage State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Organism: Nocardia seriolae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-11-13 Classification: HYDROLASE Ligands: UMP, PO4, TRS, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Cfhhacs From Chloroflexi Bacterium In The Complex With Thdp And Adp
Organism: Chloroflexi bacterium hgw-chloroflexi-9
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, ADP, MG, EDO, PEG |