Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Cryo-Em Structure Of Uropathogenic Escherichia Coli Cysk:Cdia:Trna Complex A
Organism: Escherichia coli, Escherichia coli 536
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: TOXIN Ligands: MG |
 |
Cryo-Em Structure Of Uropathogenic Escherichia Coli Cysk:Cdia:Trna Complex B
Organism: Escherichia coli, Escherichia coli 536
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: TOXIN Ligands: MG |
 |
Crystal Structure Of Catabolite Repressor Acivator From E. Coli In Complex With Hepes
Organism: Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-04-24 Classification: GENE REGULATION Ligands: EPE |
 |
Crystal Structure Of Catabolite Repressor Acivator From E. Coli In Complex With Sulisobenzone
Organism: Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: I4Y, PO4, EPE, EDO |
 |
Crystal Structure Of Catabolite Repressor Acivator From E. Coli In Complex With Hepes
Organism: Escherichia coli o6:k15:h31 (strain 536 / upec)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-12-22 Classification: GENE REGULATION Ligands: EPE, GOL |
 |
Organism: Escherichia coli o6:k15:h31 (strain 536 / upec)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-12-22 Classification: GENE REGULATION Ligands: EDO, PO4, MG |
 |
Organism: Escherichia coli o157:h7, Escherichia coli o6:k15:h31 (strain 536 / upec)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-07-27 Classification: TOXIN |
 |
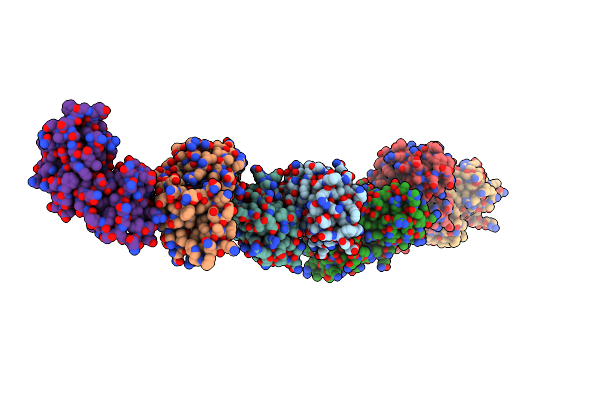
Cdia-Ct From Uropathogenic Escherichia Coli In Complex With Cognate Immunity Protein And Cysk
Organism: Escherichia coli o157:h7, Escherichia coli o6:k15:h31 (strain 536 / upec)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-07-27 Classification: TOXIN |
 |
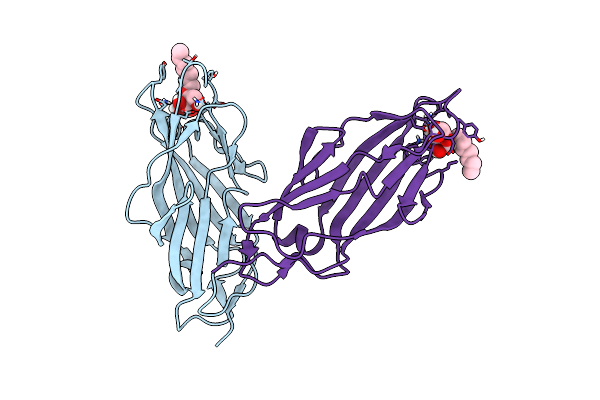
Organism: Escherichia coli o6:k15:h31 (strain 536 / upec)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-06-22 Classification: HYDROLASE |
 |
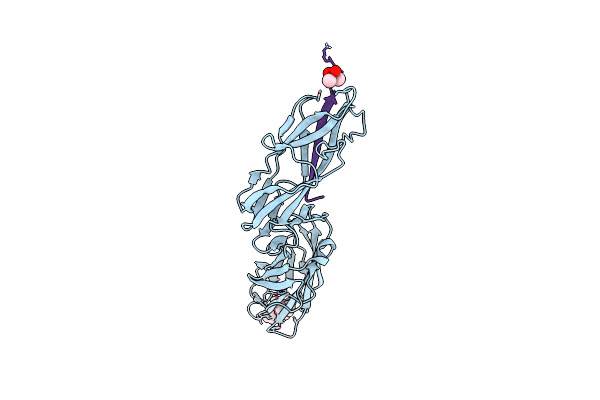
Crystal Structure Of The Fimh Lectin Domain From E.Coli F18 In Complex With Heptyl Alpha-D-Mannopyrannoside
Organism: Escherichia coli o6:k15:h31
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2016-01-27 Classification: CELL ADHESION Ligands: KGM |
 |
Crystal Structure Of A Fimh*Dsg Complex From E.Coli F18 With Bound Heptyl Alpha-D-Mannopyrannoside
Organism: Escherichia coli o6:k15:h31
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-01-27 Classification: CELL ADHESION Ligands: KGM, CAC |
 |
Nmr Structure Of The Cytidine Repressor Dna Binding Domain In Presence Of Operator Half-Site Dna
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2011-06-29 Classification: TRANSCRIPTION REGULATOR |
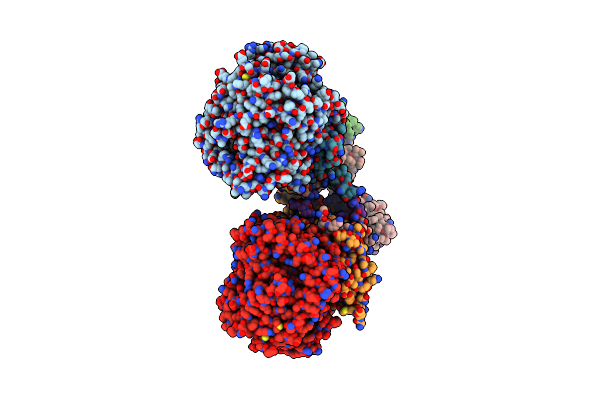 |
Crystal Structure Of Menaquinol:Oxidoreductase In Complex With Oxaloacetate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, OAA, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Fumarate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: FAD, FUM, FES, F3S, SF4 |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Escherichia coli O6:K15:H31 (strain 536 / UPEC)
Method: Alphafold Release Date: Classification: NA Ligands: NA |