Search Count: 227
 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Cleavage Intermediate 2)
Organism: Acidothermus cellulolyticus 11b, Acidothermus cellulolyticus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Cleavage Intermediate 1)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Pre-Cleavage)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
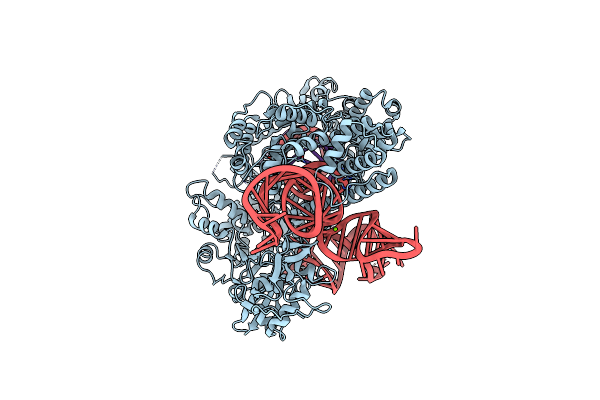
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Post-Cleavage 2)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
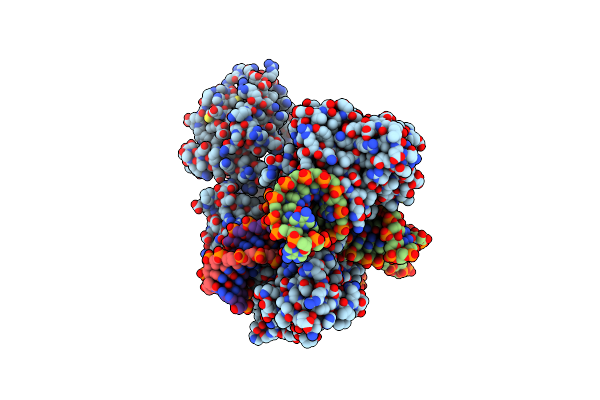
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Target Bound)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
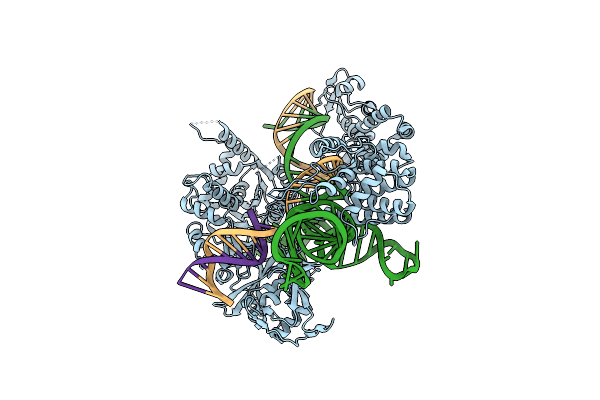
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Post-Cleavage 1)
Organism: Acidothermus cellulolyticus 11b, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Crystal Structure Of Acecas9 Bound With Guide Rna And Dna With 5'-Nnncc-3' Pam
Organism: Acidothermus cellulolyticus (strain atcc 43068 / 11b), Acidothermus cellulolyticus 11b, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2020-11-18 Classification: RNA BINDING PROTEIN/DNA/RNA |
 |
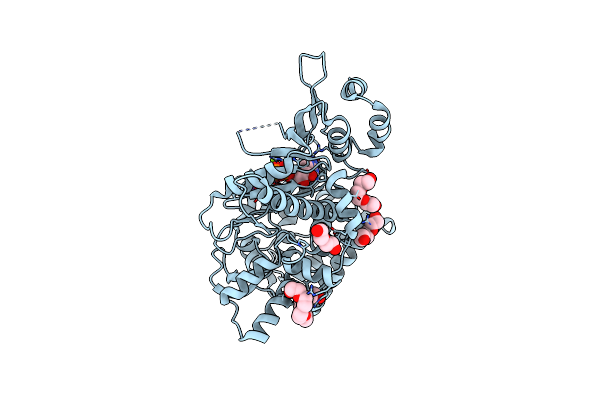
Crystal Structure Of Acecas9 Bound With Guide Rna And Dna With 5'-Nnntc-3' Pam
Organism: Acidothermus cellulolyticus (strain atcc 43068 / 11b), Acidothermus cellulolyticus 11b, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2020-11-18 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Adp And Phosphate
Organism: Acidothermus cellulolyticus (strain atcc 43068 / 11b)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: ADP, MG, PO4, P6G, PGE, PEG, NA |
 |
Crystal Structure Of Carbohydrate Transporter Acei_1806 From Acidothermus Cellulolyticus 11B, Target Efi-510965, In Complex With Myo-Inositol
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-17 Classification: TRANSPORT PROTEIN Ligands: INS, CIT |
 |
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2013-12-11 Classification: CELLULOSE BINDING PROTEIN Ligands: K, EDO, GOL, ACT, NA, FMT |
 |
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-09-12 Classification: HYDROLASE |
 |
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Atp
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-09-12 Classification: HYDROLASE Ligands: ATP, MG |
 |
Crystal Structure Of A Predicted Zincin-Like Metalloprotease (Acel_2062) From Acidothermus Cellulolyticus 11B At 1.80 A Resolution
Organism: Acidothermus cellulolyticus 11b
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-08-19 Classification: UNKNOWN FUNCTION Ligands: ACT, CA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Acidothermus cellulolyticus (strain ATCC 43068 / 11B)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

