Search Count: 488
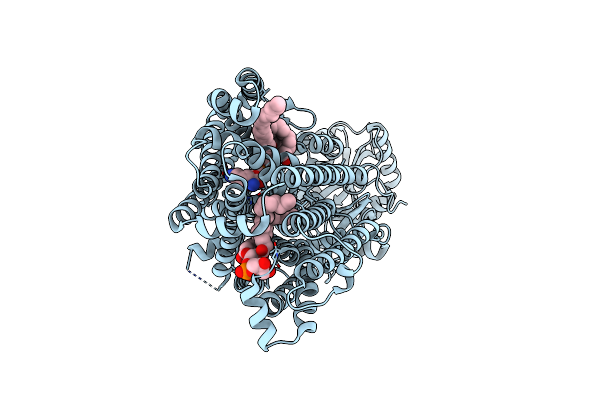 |
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: ELECTRON MICROSCOPY Resolution:2.99 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: PGW, 1K1 |
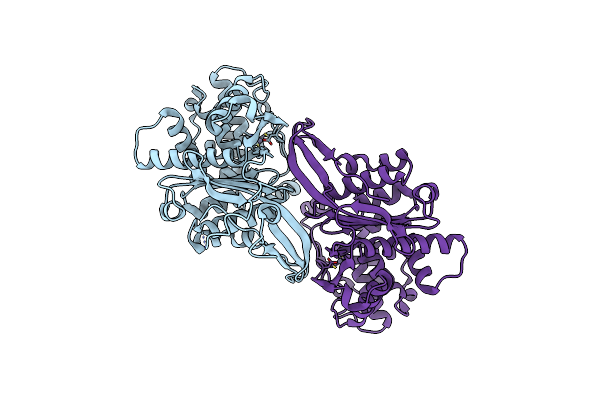 |
Crystal Structure Of Rhizobium Etli Inducible L-Asparaginase Reav Solved By S-Sad (Orthorhombic Form Start)
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: ZN, CL |
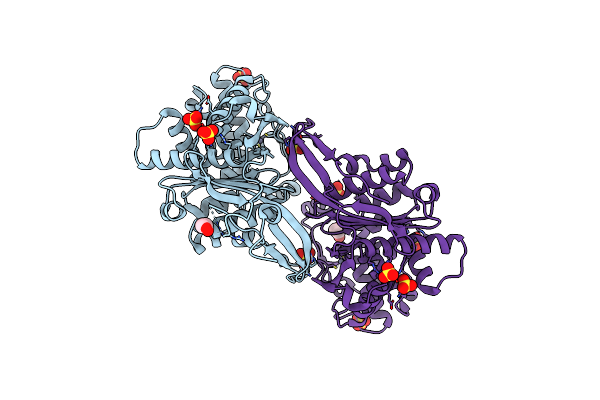 |
Crystal Structure Of Rhizobium Etli Inducible L-Asparaginase Reav (Orthorhombic Form Op)
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: ZN, EDO, CL, SO4 |
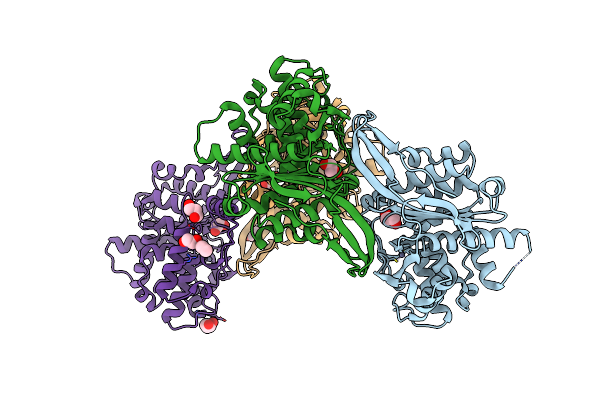 |
Crystal Structure Of Rhizobium Etli Inducible L-Asparaginase Reav (Monoclinic Form Mp1)
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: ZN, CL, EDO, GOL, EOH |
 |
Crystal Structure Of Rhizobium Etli Inducible L-Asparaginase Reav (Monoclinic Form Mp2)
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: ZN, EDO |
 |
Crystal Structure Of Rhizobium Etli Inducible L-Asparaginase Reav (Monoclinic Form Mc)
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: PEG, ZN |
 |
Crystal Structure Of Nadph-Dependent 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadph And Oxalate
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: NDP, OXD |
 |
Crystal Structure Of An Abc Solute Binding Protein From Rhizobium Etli Cfn 42 (Rhe_Pf00037,Target Efi-511357) In Complex With Alpha-D-Apiose
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-04-20 Classification: SOLUTE-BINDING PROTEIN Ligands: XXM, CA |
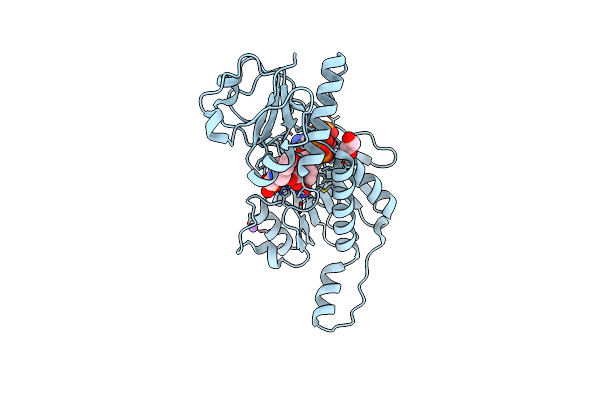 |
Probable 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadp, Hepes And L(+)-Tartaric Acid
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-06-17 Classification: OXIDOREDUCTASE Ligands: EPE, PEG, NAP, TLA, NA, CL |
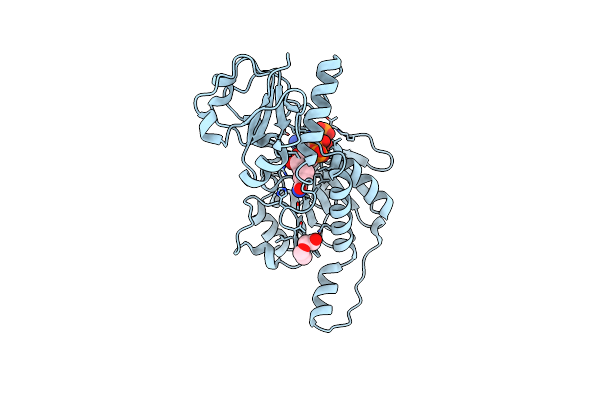 |
Probable 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadph
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-31 Classification: OXIDOREDUCTASE Ligands: NDP, CL, PEG |
 |
Crystal Structure Of Ribose Transporter Solute Binding Protein Rhe_Pf00037 From Rhizobium Etli Cfn 42, Target Efi-511357, In Complex With D-Ribose
Organism: Rhizobium etli cfn 42
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-24 Classification: TRANSPORT PROTEIN Ligands: RIP, CL, CA |
 |
Crystal Structure Of Sugar Transporter Rhe_Pf00321 From Rhizobium Etli, Target Efi-510806, An Open Conformation
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-05 Classification: TRANSPORT PROTEIN Ligands: CL, ACT |
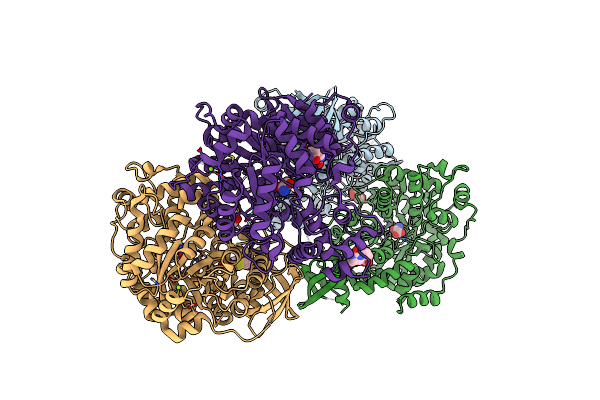 |
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With Oxamate And Biotin
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-09-10 Classification: LIGASE Ligands: ZN, MG, CL, BTN, OXM, GOL |
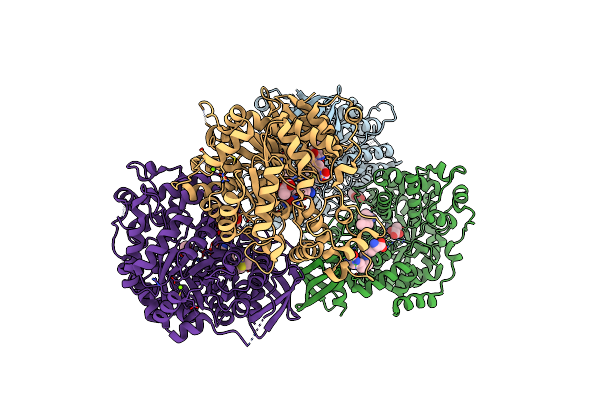 |
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With Pyruvate And Biocytin
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-09-10 Classification: LIGASE Ligands: ZN, MG, CL, PYR, BYT, GOL |
 |
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With 3-Bromopyruvate
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-08-13 Classification: LIGASE Ligands: ZN, BPV, CL, MG, GOL |
 |
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With Oxalate
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: ZN, OXL, MG, CL, GOL |
 |
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With 3-Hydroxypyruvate
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: ZN, 3PY, BTN, MG, CL, GOL |
 |
Crystal Structure Of Probable Sugar Kinase Protein From Rhizobium Etli Cfn 42 Complexed With Adenosine
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-07-03 Classification: TRANSFERASE Ligands: ADN, DMS, K |
 |
Crystal Structure Of Probable Sugar Kinase Protein From Rhizobium Etli Cfn 42 Complexed With Cytidine
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-03 Classification: TRANSFERASE Ligands: ADN, CTN, DMS, K |
 |
Crystal Structure Of Probable Sugar Kinase Protein From Rhizobium Etli Cfn 42 Complexed With Guanosine
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-03 Classification: TRANSFERASE Ligands: ADN, GMP, DMS, K |

