Search Count: 1,343
 |
Organism: Ixodes ricinus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: SO4, E64 |
 |
Organism: Xanthomonas citri
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 12C, SO4 |
 |
Organism: Klebsiella michiganensis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LYASE Ligands: ANP, MG, A1H6S, MES, CL, S2G |
 |
Organism: Streptomyces niveus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: PO4, FE |
 |
Bifunctional Non-Haem Iron Oxygenase Novr Involved In Novobiocin Biosynthesis With Product Bound
Organism: Streptomyces niveus
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: LYASE Ligands: A1JKI, FE, SO4, EDO |
 |
The Structure Of A Family 168 Glycoside Hydrolase From The Marine Bacterium Muricauda Eckloniae.
Organism: Flagellimonas eckloniae
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Tyrosine Decarboxylase H203F Mutant In Complex With The Cofactor Plp And Tyrosine
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: TYR |
 |
Crystal Structure Of Tyrosine Decarboxylase In Complex With The Cofactor Plp And Inhibitor Carbidopa
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: PLP, 142 |
 |
Crystal Structure Of The Transpeptidase Domain Of Pbp2 From The Neisseria Gonorrhoeae Cephalosporin Decreased Susceptibility Strain 35/02 In Complex With Boronate Inhibitor Vnrx-6884
Organism: Neisseria gonorrhoeae 35/02
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-22 Classification: LIGASE Ligands: A1BJC |
 |
Crystal Structure Of The Transpeptidase Domain Of Pbp2 From The Neisseria Gonorrhoeae Cephalosporin Decreased Susceptibility Strain 35/02 In Complex With Boronate Inhibitor Vnrx-6752
Organism: Neisseria gonorrhoeae 35/02
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-01-22 Classification: LIGASE Ligands: A1BJB |
 |
Organism: Mycoplasmoides pneumoniae 19294
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP |
 |
Organism: Mycoplasmoides pneumoniae 19294
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
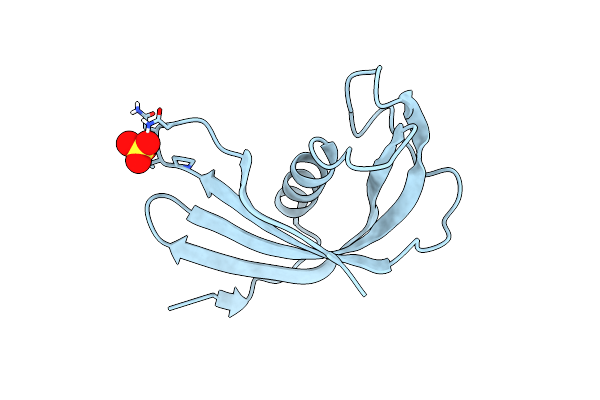 |
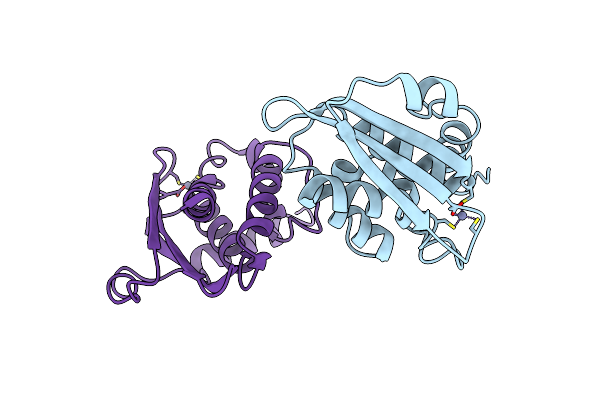
Crystal Structure Of Amacstatin-1, A Cystatin From The Hard Tick Amblyomma Maculatum
Organism: Amblyomma maculatum
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-11-13 Classification: PROTEIN BINDING Ligands: SO4 |
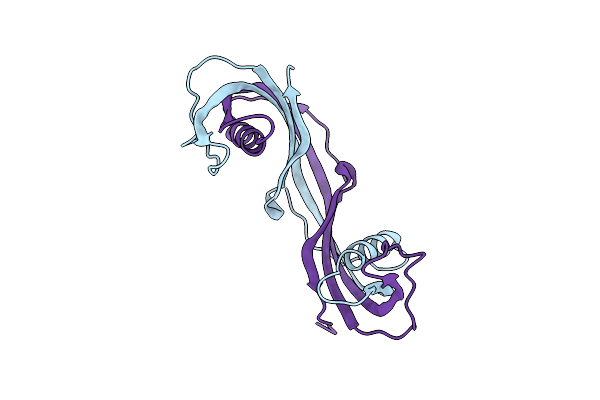 |
Crystal Structure Of Domain Swapped Dimer Of Amacstatin-2, A Cystatin From The Hard Tick Amblyomma Maculatum
Organism: Amblyomma maculatum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-11-13 Classification: PROTEIN BINDING |
 |
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-23 Classification: PLANT PROTEIN |
 |
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-10-23 Classification: PLANT PROTEIN |
 |
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-10-23 Classification: PLANT PROTEIN Ligands: EV1 |
 |
Organism: Streptomyces aureus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Arylamine N-Acyltransferase From Streptomyces Aureus Complexed With Acyl-Snac
Organism: Streptomyces aureus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: I7G, SO4 |
 |
Organism: Norovirus hu/gi.4/s50/2008/lilla edet/sweden
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN |

