Search Count: 156
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, ACT, GOL |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With A Nanobody
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: GOL, NAG |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN |
 |
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: ACT, IMD, GOL, SO4, NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: IMD, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO, MAN |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: PLANT PROTEIN Ligands: EDO, IMD |
 |
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-11-06 Classification: PLANT PROTEIN Ligands: PEG, BME, ACT |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-02-28 Classification: PLANT PROTEIN Ligands: SO4, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-02-21 Classification: TRANSFERASE |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-11-10 Classification: PLANT PROTEIN Ligands: NAG |
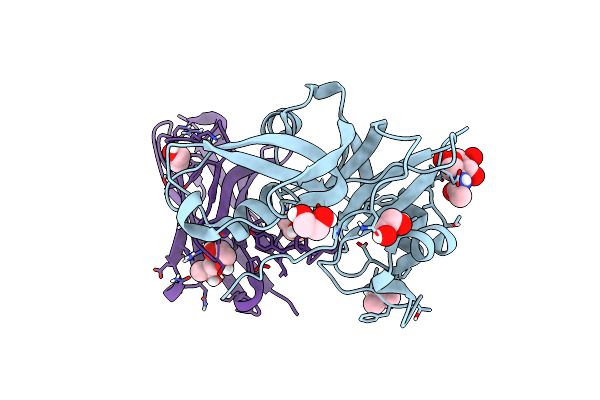 |
Structural Signatures In Epr3 Define A Unique Class Of Plant Carbohydrate Receptors
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-08-05 Classification: PLANT PROTEIN Ligands: NAG, IPA, EDO |
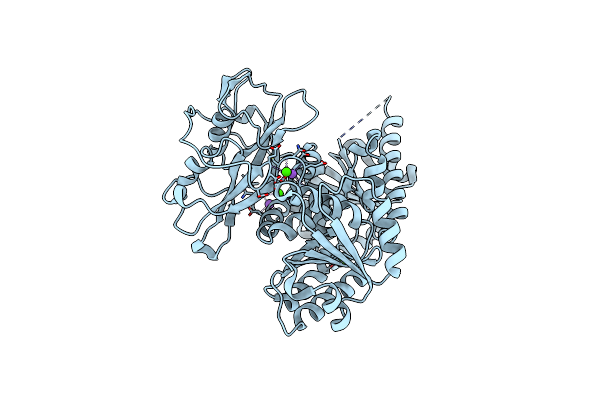 |
Crystal Structure Of The Ljcastor Gating Ring In The Ca2+ And Na+ Condition
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: CA, MG, NA |
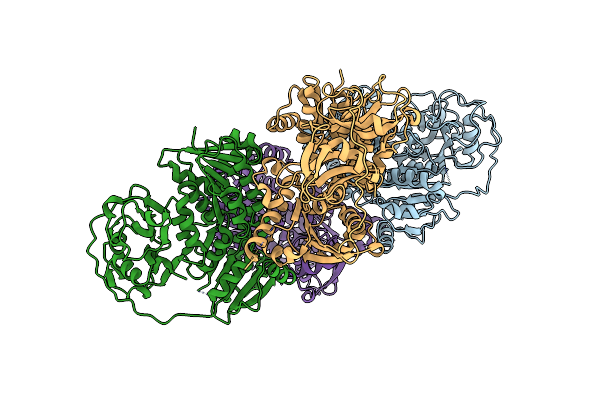 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: CA, MG, K, NA |
 |
Receptor Mediated Chitin Perception In Legumes Is Functionally Seperable From Nod Factor Perception
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-23 Classification: PLANT PROTEIN Ligands: SO4 |
 |
 |

