Search Count: 40,765
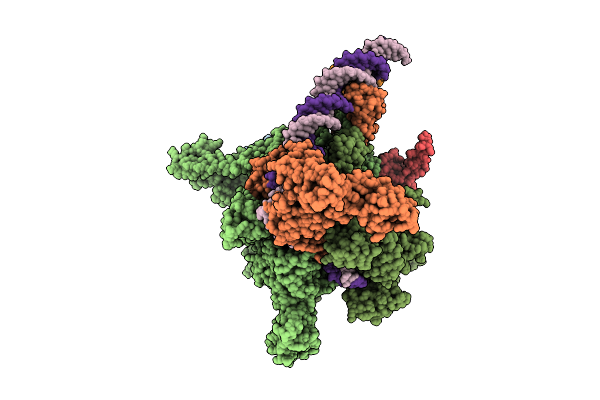 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
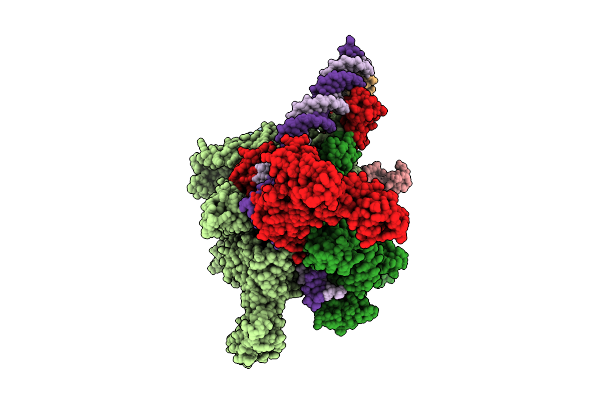 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
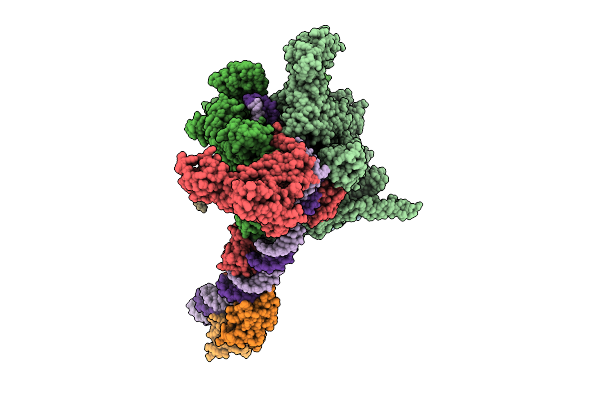 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
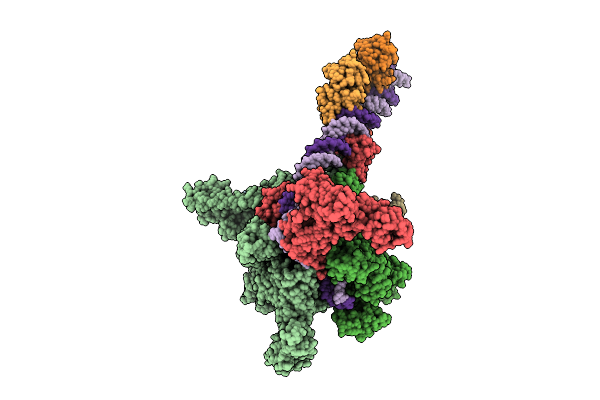 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
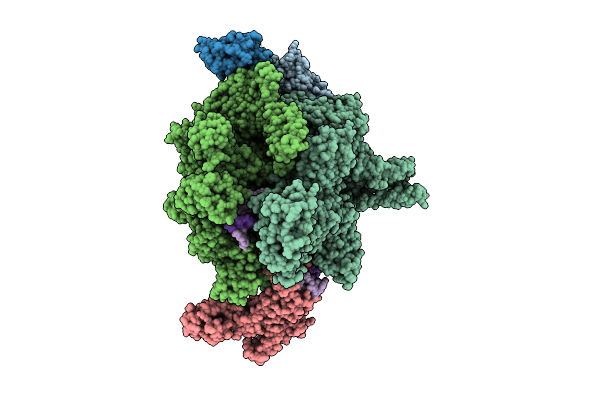 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
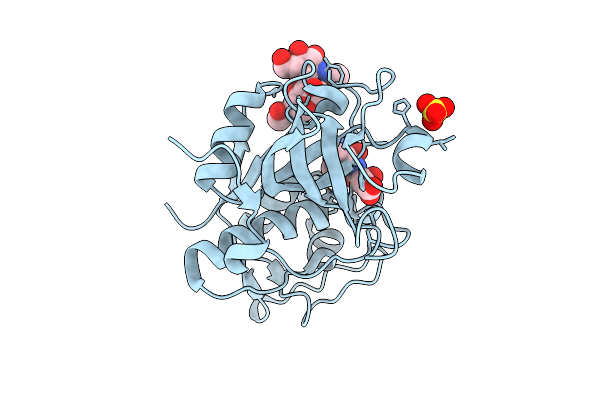 |
Organism: Ixodes ricinus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: SO4, E64 |
 |
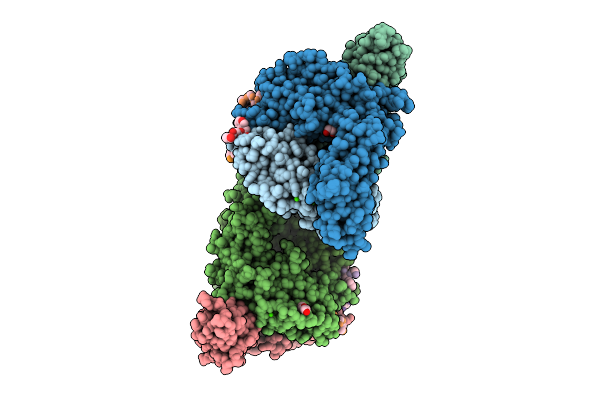
Organism: Finegoldia magna atcc 29328, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE/DNA Ligands: ANP, SF4 |
 |
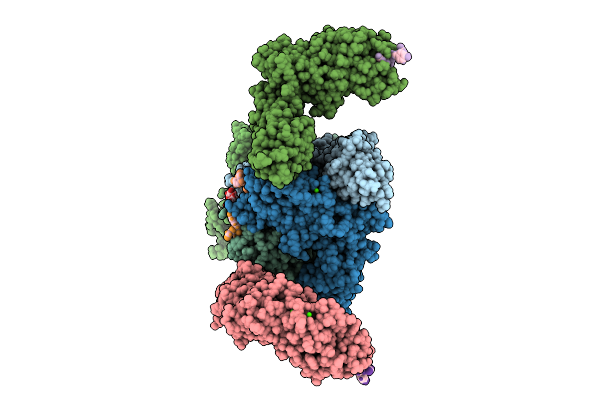
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agwtd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA, PEG, EDO |
 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agytd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Organism: Xanthomonas citri
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 12C, SO4 |
 |
Organism: Homo sapiens, Sars bat coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RIBOSOME |
 |
Organism: Homo sapiens, Sars bat coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RIBOSOME |
 |
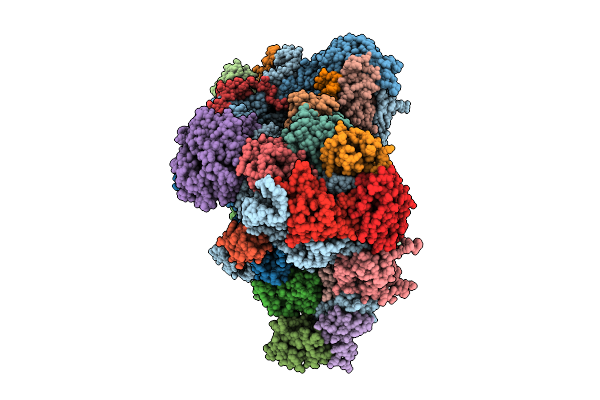
Organism: Homo sapiens, Sars bat coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RIBOSOME |
 |
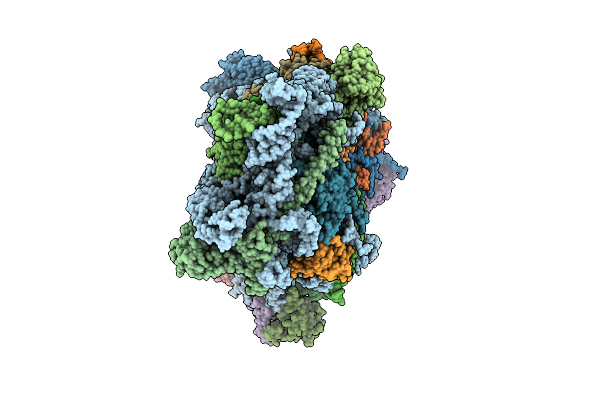
Organism: Homo sapiens, Sars bat coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RIBOSOME |
 |
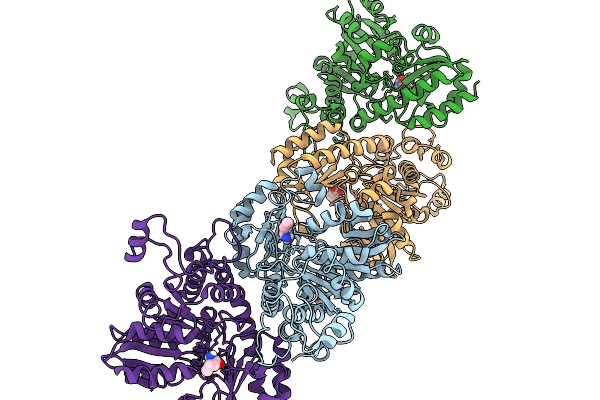
Organism: Homo sapiens, Sars bat coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RIBOSOME |
 |
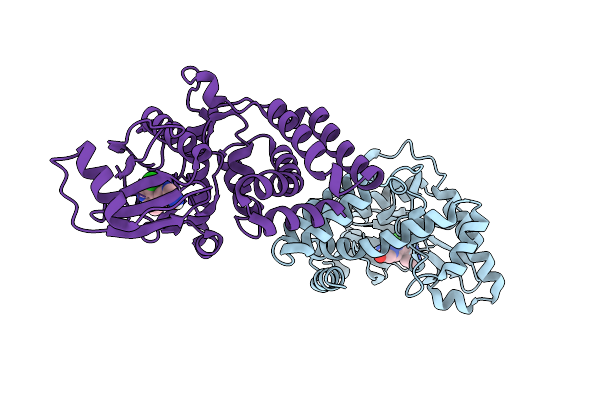
Crystal Structure Of Highly Stable Methionine Gamma-Lyase From Thermobrachium Celere In Complex With Plp And Norleucine
Organism: Thermobrachium celere
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: NLE, EDO |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ANTIBIOTIC Ligands: DMS, A1I8X |
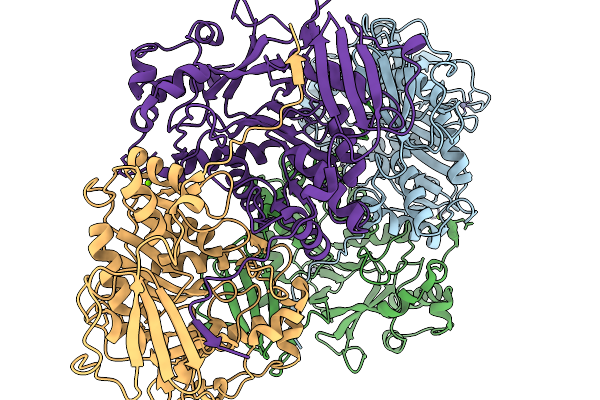 |
Organism: Clostridium cavendishii
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIGASE Ligands: ZN, MG |
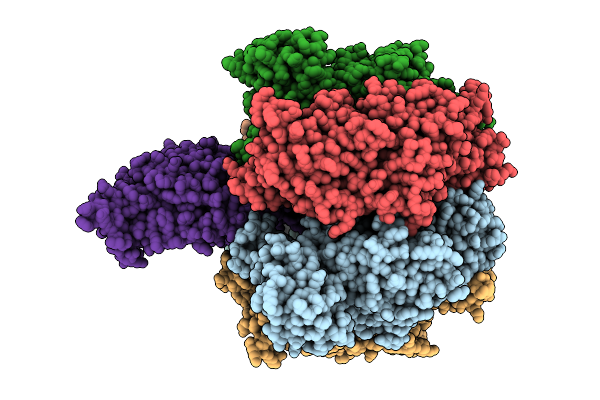 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: ELECTRON TRANSPORT Ligands: MG, ATP, PO4, ADP |
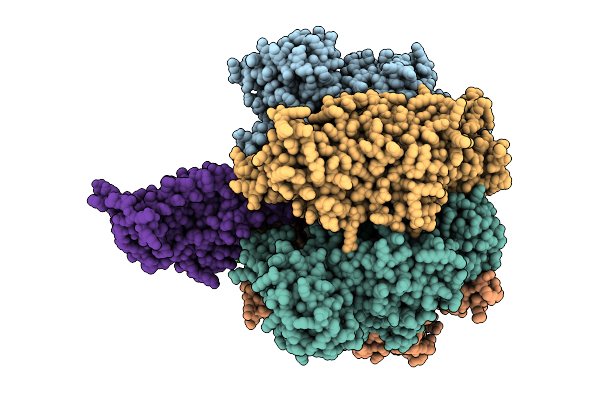 |
Organism: Bacillus sp. ps3, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |

