Search Count: 1,442
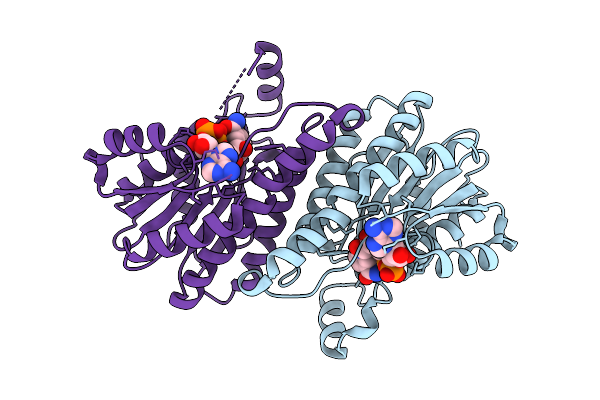 |
Crystal Structure Of L-2-Keto-3-Deoxypentonate 4-Dehydrogenase Bound To Nad(H)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD |
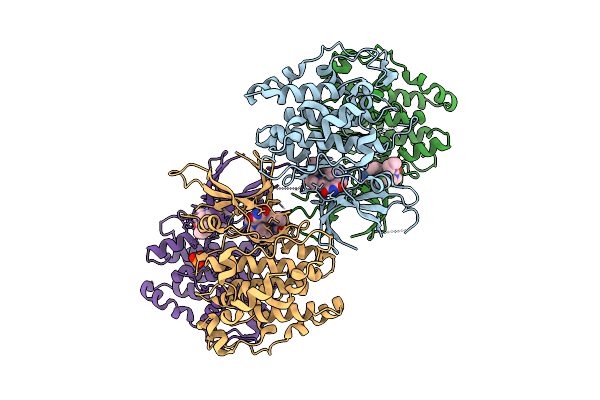 |
Toxoplasma Gondii Gsk3B Bound To Ly2090314 And Disulphide Bonded Through The C223 Residue
Organism: Toxoplasma gondii rh
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: A1IYF, PO4 |
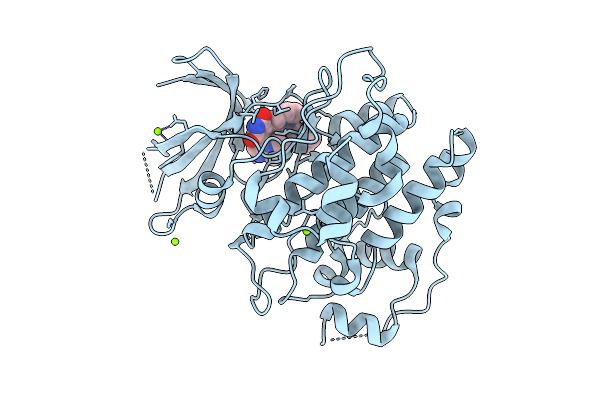 |
Organism: Toxoplasma gondii rh
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: A1IYF, MG |
 |
Crysral Structure Of 2-Keto-3-Deoxypentonate 4-Dehydrogenase From Herbaspirillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE |
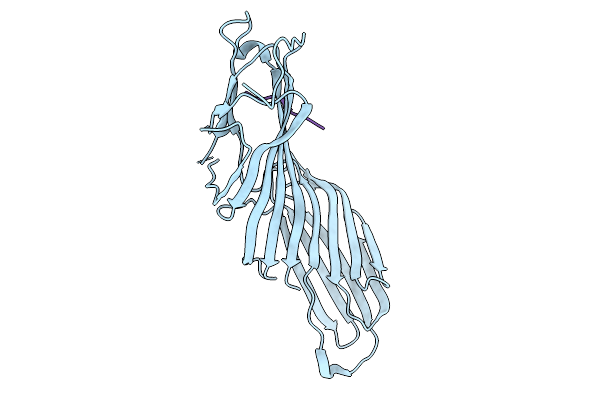 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-39)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-32; S31E And S32E)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-39; S31E And S32E)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Organism: Hepatitis e virus (strain pakistan), Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
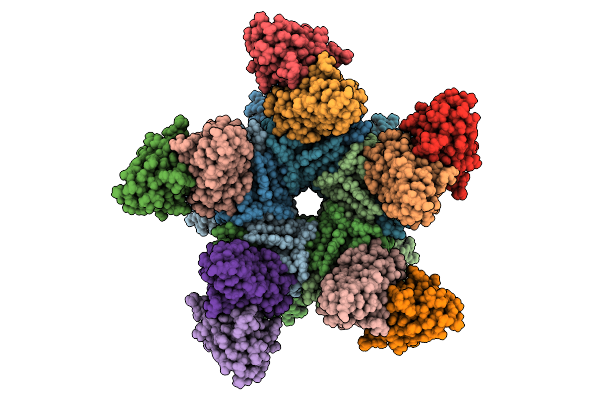
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM |
 |
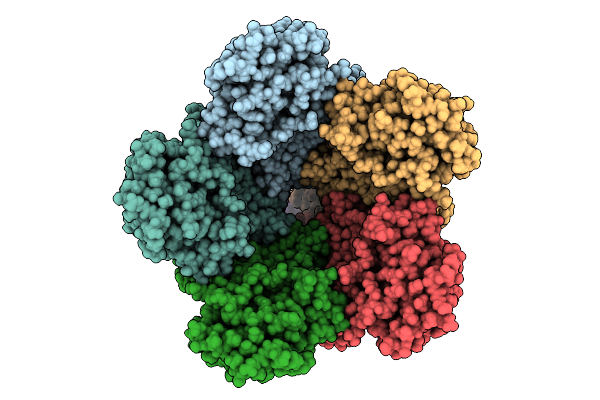
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM |
 |
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM |
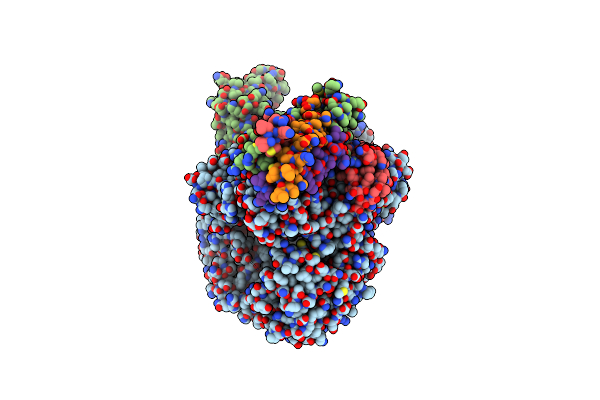 |
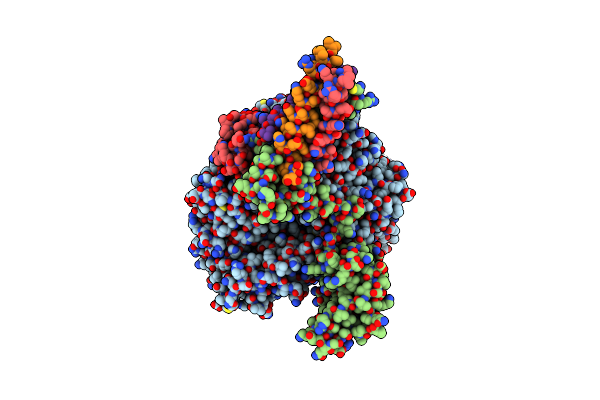
Cryo-Em Structure Of The Rna-Dependent Rna Polymerase Complex From Marburg Virus
Organism: Marburg virus - musoke, kenya, 1980, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of The Rna-Dependent Rna Polymerase Complex From Marburg Virus
Organism: Marburg virus - musoke, kenya, 1980, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Alphapapillomavirus 10
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human papillomavirus type 16
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human papillomavirus 18
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Streptococcus sp. group g, Human papillomavirus 16
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: LIGASE |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Target-Cleaved State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN Ligands: MN |

