Search Count: 341
 |
Crystal Structure Of The Pcryo_0617 Oxidoreductase/Decarboxylase From Psychrobacter Cryohalolentis K5 In The Presence Of Nad And Udp
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: OXIDOREDUCTASE Ligands: NAD, UDP, NA, MES |
 |
Crystal Structure Of The Pcryo_0618 Aminotransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Its Internal Aldimine
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: EDO, NA, A1ADI, CL |
 |
Crystal Structure Of The Pcryo_0618 Aminotransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Pmp And Glutamate
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: PMP, A1ADI, CL, EDO, MG, PDG, NA |
 |
Crystal Structure Of The Pcryo_0619 N-Acetryltransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Coa-Disulfide
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: 5NG |
 |
Crystal Structure Of The Pcryo_0619 N-Acetyltransferase From Psychrobacter Cryohalolentis K5 Int He Presence Of Acetyl Coenzyme A
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: ACO, COA, EDO, EP1, PAP |
 |
Crystal Structure Of The Pcryo_0619 N-Acetyltransferase From Psychrobacter Cryohalolentis K5
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SO4, MPD, EDO |
 |
Structure Of Pcryo_0615 From Psychrobacter Cryohalolentis, An N-Acetyltransferase Required To Produce Diacetamido-2,3-Dideoxy-D-Glucuronic Acid
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: MJZ, COA, NA |
 |
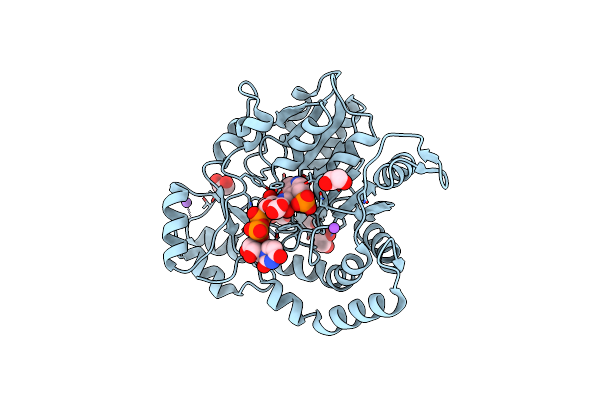
Crystal Structure Of Pcryo_0616, The Aminotransferase Required To Synthesize Udp-N-Acetyl-3-Amino-D-Glucosaminuronic Acid (Udp-Glcnac3Na)
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-11-23 Classification: TRANSFERASE |
 |
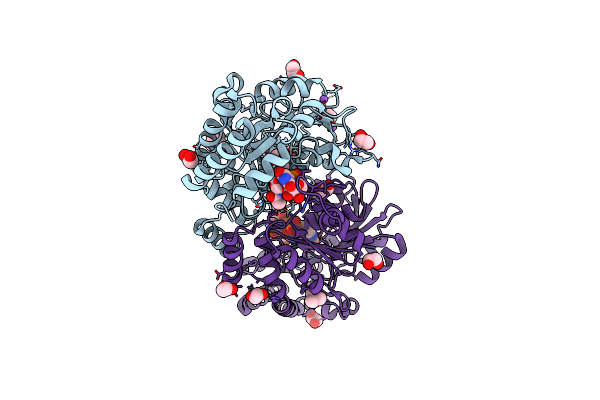
Rystal Structure Of Pcryo_0616, The Aminotransferase Required To Synthesize Udp-N-Acetyl-3-Amino-D-Glucosaminuronic Acid (Udp-Glcnac3Na), Incomplete With Its External Aldimine Reaction Intermediate
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: ULP, EDO, NA |
 |
X-Ray Structure Of The Pcryo_0638 Aminotransferase From Psychrobacter Cryohalolentis
Organism: Psychrobacter cryohalolentis (strain atcc baa-1226 / dsm 17306 / vkm b-2378 / k5)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: PMP, 4RA, EDO, TMA, NA |
 |
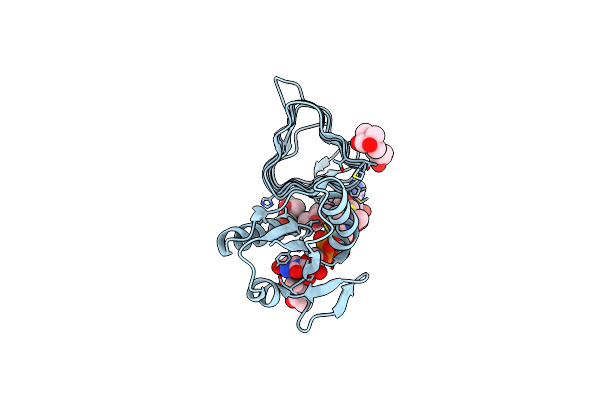
X-Ray Structure Of The N-Acetyltransferase Pcryo_0637 From Psychrobacter Cryohalolentis In The Presence Of Udp And Acetyl-Conezyme A
Organism: Psychrobacter cryohalolentis (strain atcc baa-1226 / dsm 17306 / vkm b-2378 / k5)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: UDP, ACO, MPD, CL |
 |
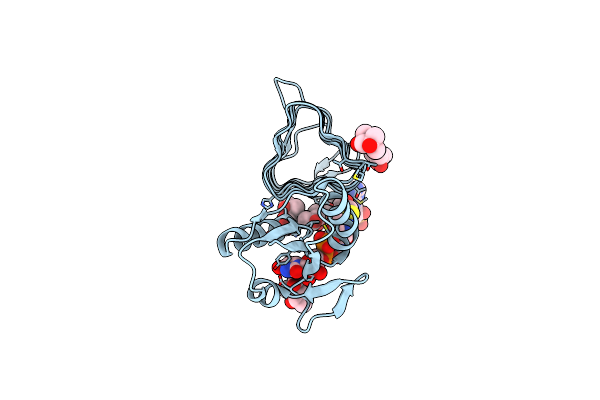
X-Ray Structure Of The N-Acetyltransferase Pcryo_0637 From Psychrobacter Cryohalolentis In The Presence Of Coenzyme A And Udp-Di-N-Acetyl-Bacillosamine
Organism: Psychrobacter cryohalolentis (strain atcc baa-1226 / dsm 17306 / vkm b-2378 / k5)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: COA, XQA, CL, MPD |
 |
X-Ray Structure Of The Psychrobacter Cryohalolentis N-Acetyltransferase Pcryo_0637 In The Presence Of Coenzyme A And
Organism: Psychrobacter cryohalolentis (strain atcc baa-1226 / dsm 17306 / vkm b-2378 / k5)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: UD4, COA, CL, MPD |
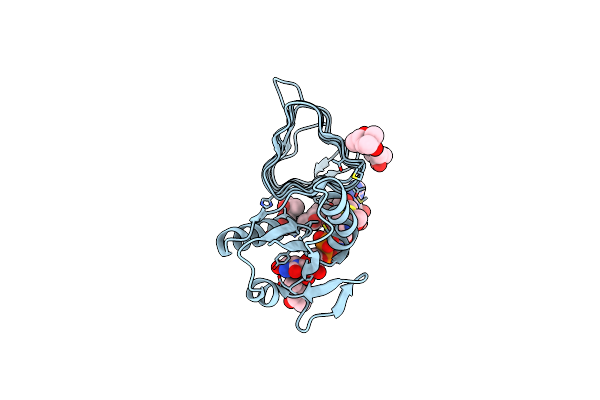 |
X-Ray Structure Of The Psychrobacter Cryohalolentis Pcryo_0637 N-Acetyltransferase In The Presene Of Its Reaction Tetrahedral Intermediate
Organism: Psychrobacter cryohalolentis (strain atcc baa-1226 / dsm 17306 / vkm b-2378 / k5)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: XQD, CL, MPD |
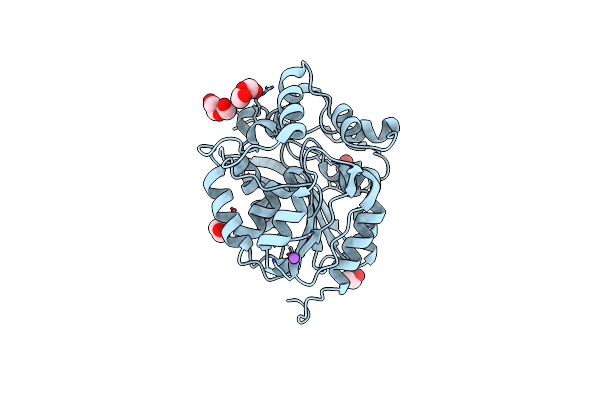 |
Crystal Structure Of Cold-Adapted Haloalkane Dehalogenase Dpca From Psychrobacter Cryohalolentis K5
Organism: Psychrobacter cryohalolentis (strain k5)
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: CL, PEG, NA |
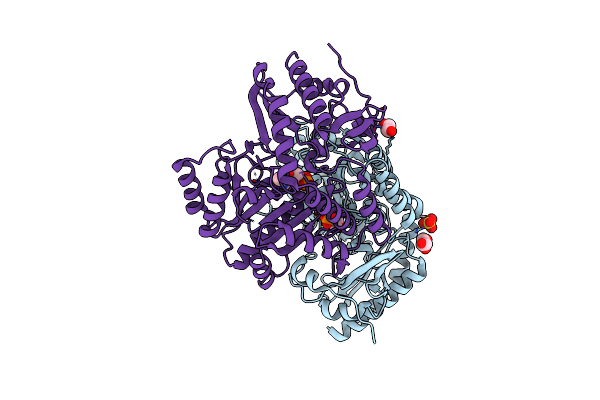 |
Crystal Structure Of Diaminopelargonic Acid Aminotransferase From Psychrobacter Cryohalolentis
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-09-26 Classification: TRANSFERASE Ligands: SO4, EDO, GOL, PLP |
 |
Crystal Structure Of Cold-Active Estarase From Psychrobacter Cryohalolentis K5T
Organism: Psychrobacter cryohalolentis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-01-07 Classification: HYDROLASE |
 |
NA
Organism: Psychrobacter cryohalolentis (strain ATCC BAA-1226 / DSM 17306 / VKM B-2378 / K5)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Psychrobacter cryohalolentis (strain ATCC BAA-1226 / DSM 17306 / VKM B-2378 / K5)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Psychrobacter cryohalolentis (strain ATCC BAA-1226 / DSM 17306 / VKM B-2378 / K5)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

