Search Count: 10
 |
The Crystal Structure Of Vypal2-C214A, A Dead Mutant Of Vypal2 Peptide Asparaginyl Ligase In Form Ii
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: PLANT PROTEIN Ligands: NAG |
 |
The Crystal Structure Of Vypal2-I244V, A More Efficient Mutant Of Vypal2 Peptide Asparaginyl Ligase In Its Active Enzyme Form
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN Ligands: NAG, EDO, GOL |
 |
The Crystal Structure Of Vypal2-C214A, A Dead Mutant Of Vypal2 Peptide Asparaginyl Ligase In Form I
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN |
 |
The Crystal Structure Of Vypal2 Peptide Asparaginyl Ligase In Its Active Enzyme Form
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-29 Classification: PLANT PROTEIN Ligands: NAG, EDO |
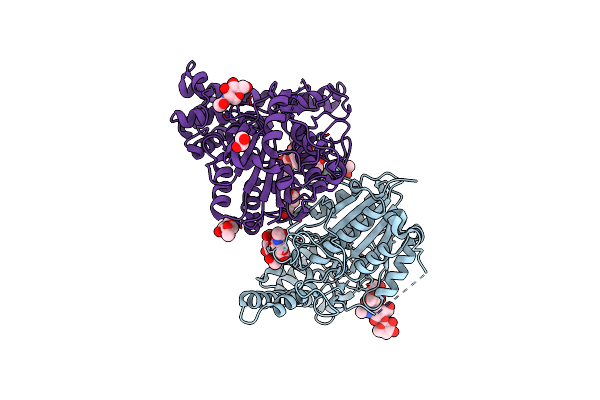 |
Organism: Viola philippica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-05-15 Classification: LIGASE Ligands: NAG, EDO, PEG |
 |
Structure Of The Core Ectodomain Of The Hepatitis C Virus Envelope Glycoprotein 2
Organism: Hepatitis c virus subtype 2a, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-12-17 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG, ARF |
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|

