Search Count: 171
 |
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN |
 |
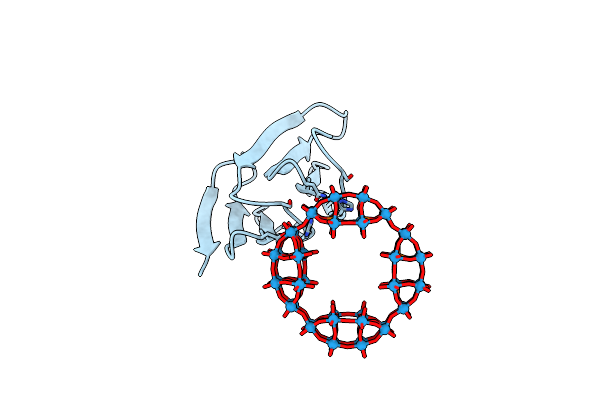
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: IR0 |
 |
Organism: Dictyoglomus thermophilum h-6-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: IR0 |
 |
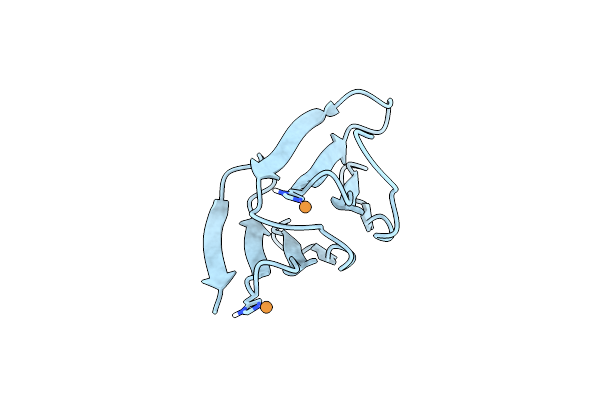
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: CD |
 |
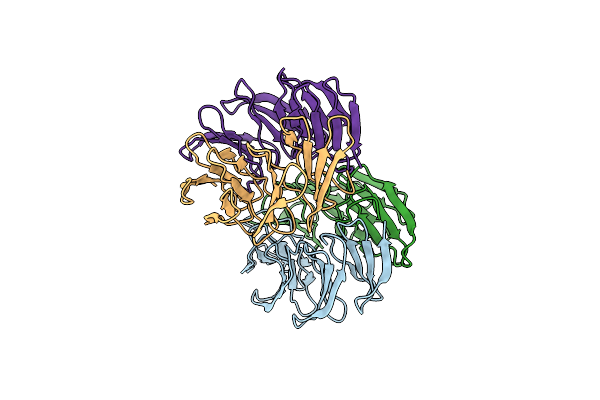
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: CU |
 |
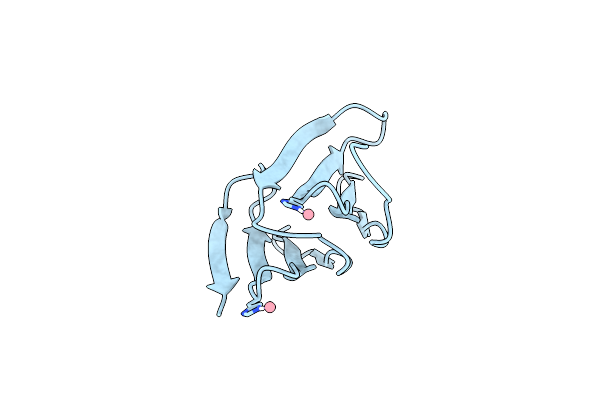
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Dictyoglomus thermophilum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: CO |
 |
Crystal Structure Of Beta-D-Xylosidase From Dictyoglomus Thermophilum In Ligand-Free Form
Organism: Dictyoglomus thermophilum h-6-12
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CIT, EDO, SO4 |
 |
Crystal Structure Of Beta-D-Xylosidase From Dictyoglomus Thermophilum Bound To Beta-D-Xylopyranose
Organism: Dictyoglomus thermophilum h-6-12
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: EDO, CIT, SO4, XYP, NA |
 |
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: TRS, MPD, CL |
 |
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: PGE, AE3 |
 |
Organism: Dictyoglomus thermophilum h-6-12
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-04-12 Classification: HYDROLASE Ligands: BMA |
 |
NA
Organism: Dictyoglomus thermophilum (strain ATCC 35947 / DSM 3960 / H-6-12)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Dictyoglomus thermophilum (strain ATCC 35947 / DSM 3960 / H-6-12)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Dictyoglomus thermophilum (strain ATCC 35947 / DSM 3960 / H-6-12)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Dictyoglomus thermophilum (strain ATCC 35947 / DSM 3960 / H-6-12)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

