Search Count: 2,441
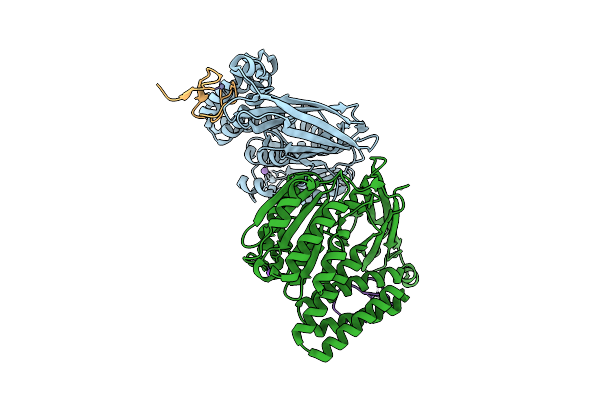 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PROTEIN BINDING |
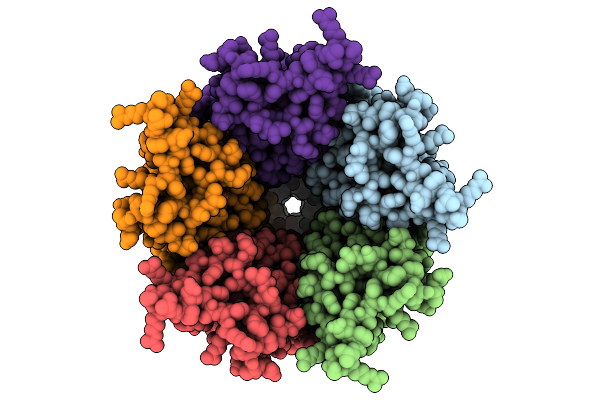 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiii Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
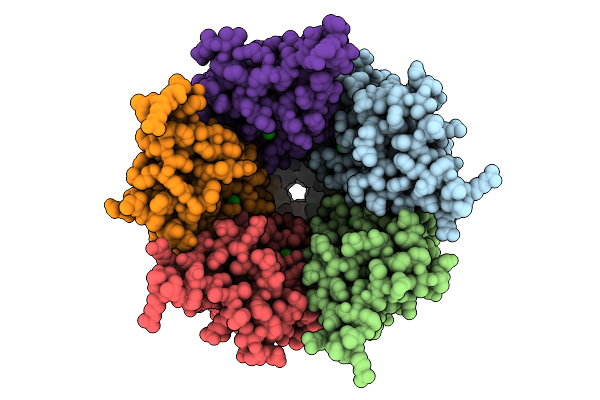 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
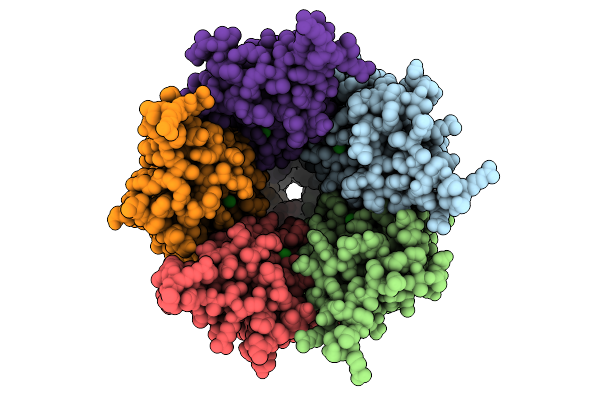 |
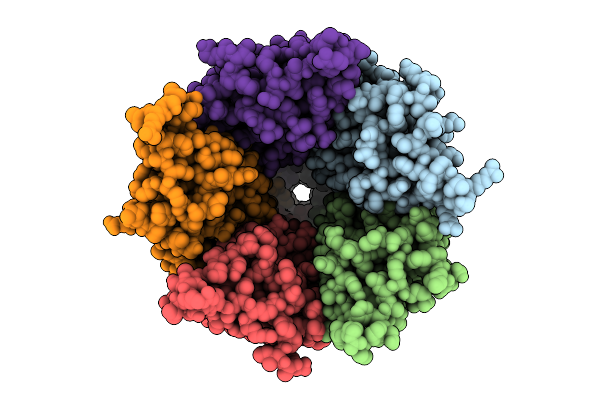
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
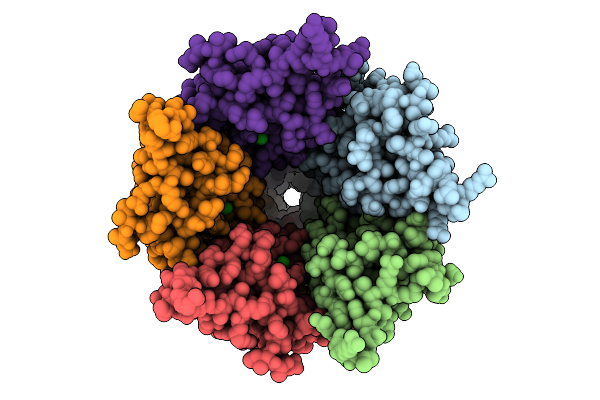 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
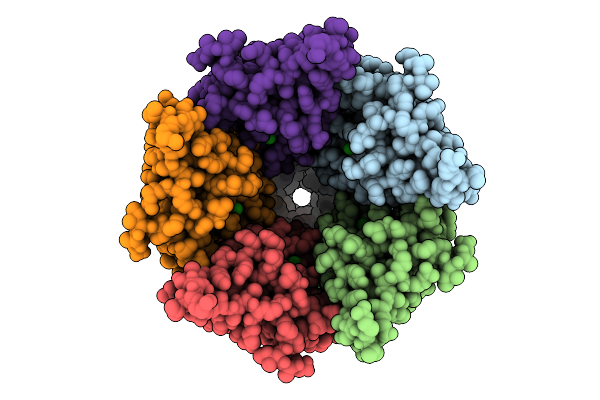 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
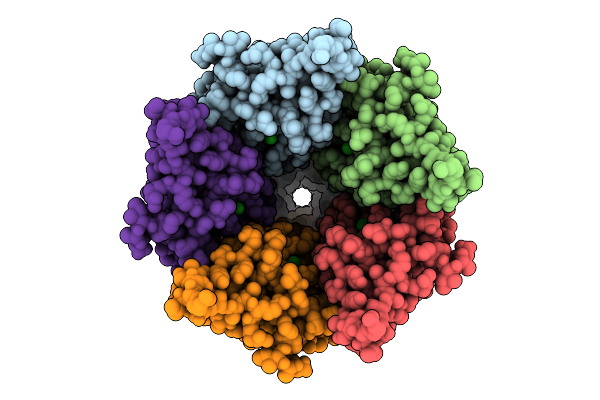 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ooooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
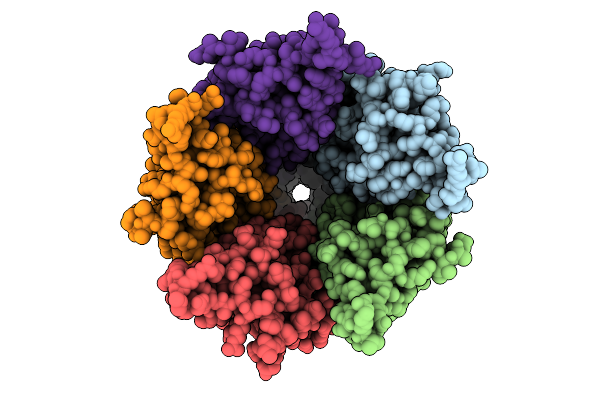
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iioio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
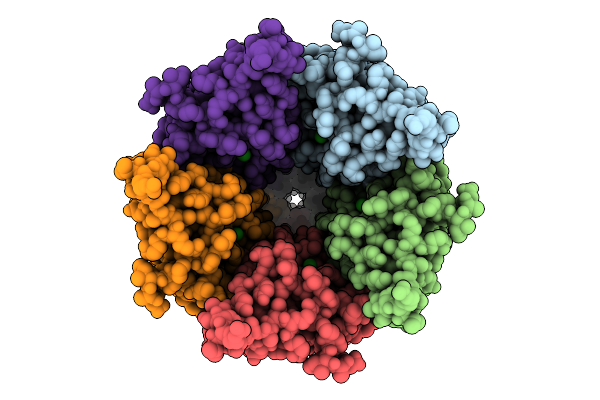
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
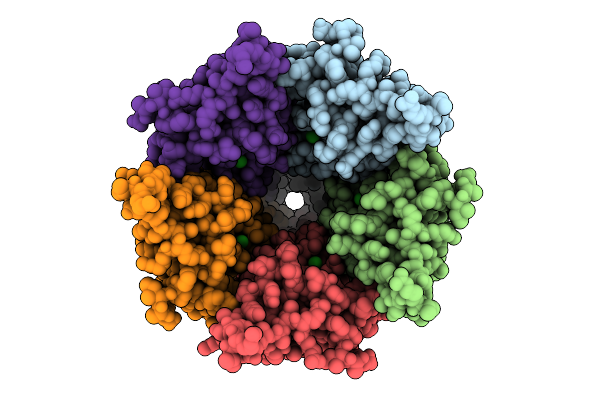
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic F116A Mutant At Ph 2.5
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic Y251A Mutant At Ph 2.5 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
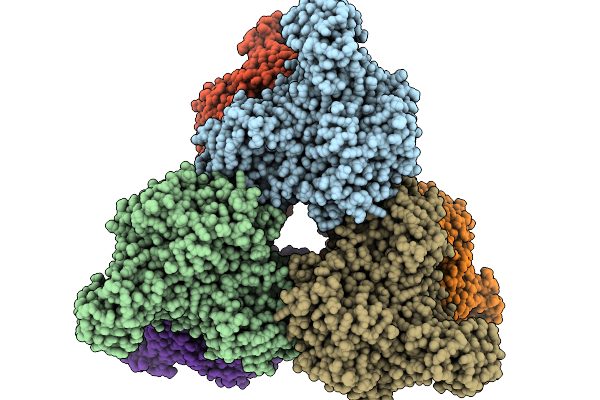
Organism: Cyclocybe aegerita
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, MG, NAG |
 |
Organism: Klebsiella aerogenes kctc 2190
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: HYDROLASE |
 |
Structure Of A Mutant (Pada-I) Unspecific Peroxygenase From Agrocybe Aegerita
Organism: Cyclocybe aegerita
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: METAL BINDING PROTEIN Ligands: MG, NAG, HEM |
 |
Structure Of Mutant Evolved From Pada-I, Unspecific Peroxygenase From Agrocybe Aegerita
Organism: Cyclocybe aegerita
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: METAL BINDING PROTEIN Ligands: MG, NAG, HEM |
 |
Organism: Homo sapiens, Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: PROTEIN BINDING |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Structure Of Saccharomyces Cerevisiae Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sah
Organism: Saccharomyces cerevisiae yjm789
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SAH, EDO, ACT |
 |
Structure Of Saccharomyces Cerevisiae Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin And Gtp
Organism: Saccharomyces cerevisiae yjm789
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, GTP, PEG, EDO, SO4, 1PE, CL |

