Search Count: 5,937
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: GOL, PEG, NA |
 |
Crystal Structure Of C43S Terpredoxin, A [2Fe-2S] Ferredoxin From Pseudomonas Sp
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-24 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Organism: Orthonairovirus songlingense
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-11-19 Classification: VIRAL PROTEIN Ligands: PEG, CL, K |
 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN |
 |
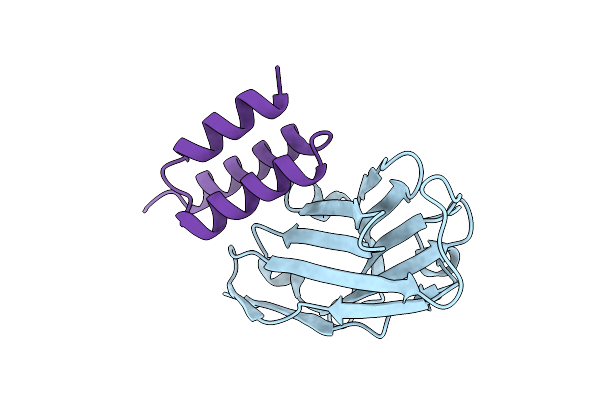
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
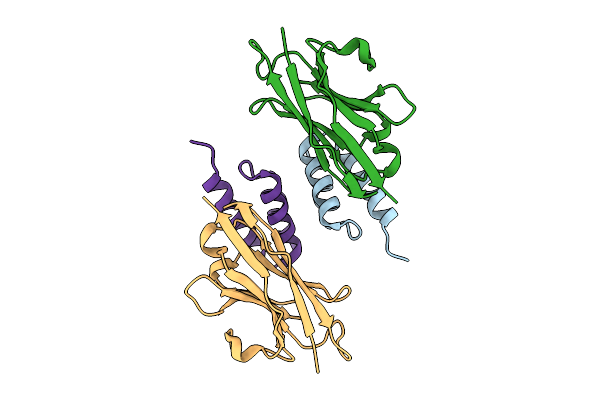
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
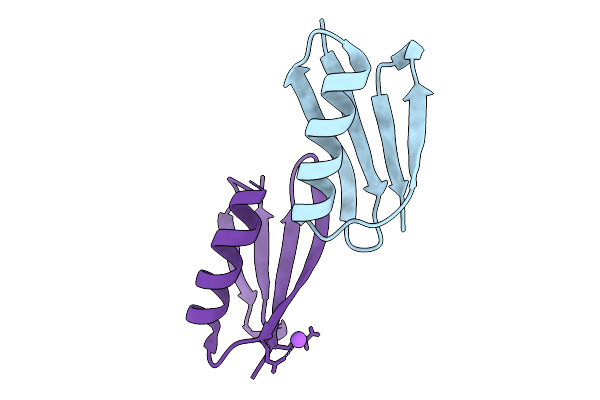
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Hsv-1 Origin Binding Protein In Complex With Double-Stranded Dna Recognition Sequence Oris With 6 Basepairs Removed From The At-Rich Region
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.77 Å Release Date: 2025-10-22 Classification: DNA BINDING PROTEIN |
 |
Hsv-1 Origin Binding Protein In Complex With Double-Stranded Dna Recognition Sequence Oris And Non-Hydrolyzable Atp Analog
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.53 Å Release Date: 2025-10-22 Classification: DNA BINDING PROTEIN Ligands: AGS, MG |
 |
Crystal Structure Of Klebsiella Pneumoniae Enoyl-Acyl Carrier Protein Reductase (Fabi) In Complex With Triclosan
Organism: Klebsiella pneumoniae subsp. pneumoniae 12-3578
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: TCL, NAD, GOL, EDO, SO4 |
 |
Solution Structure Of Staphylococcus Aureus Response Regulator Arlr Dna-Binding Domain
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN |
 |
X-Ray Structure Of The B1 Domain Of Streptococcal Protein G Triple Mutant T2Q, N8D, And N37D (Gb1-Qdd).
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2025-09-03 Classification: IMMUNE SYSTEM |
 |
Organism: Horseshoe bat sarbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
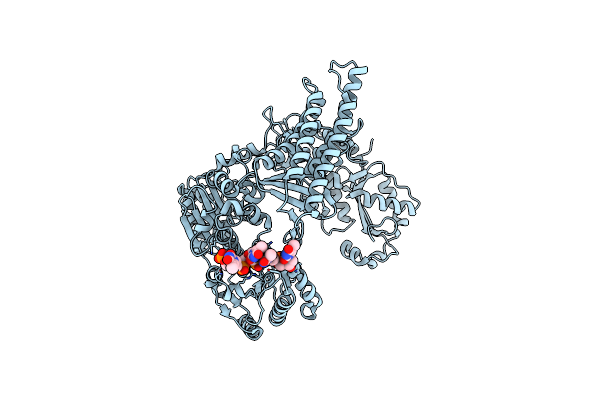 |
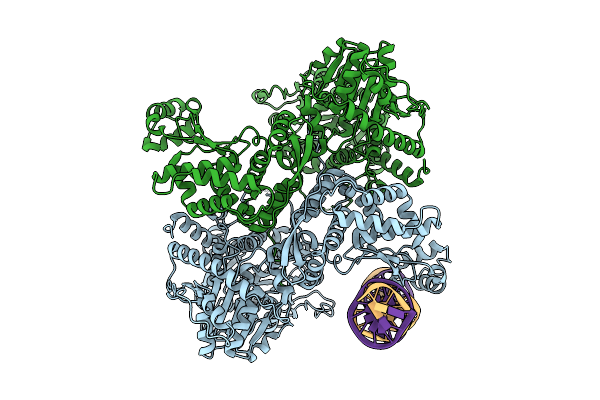
Crystal Structure Of Staphylococcus Aureus Ding Protein In Complex With Ssdna
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2025-06-25 Classification: HYDROLASE/DNA |
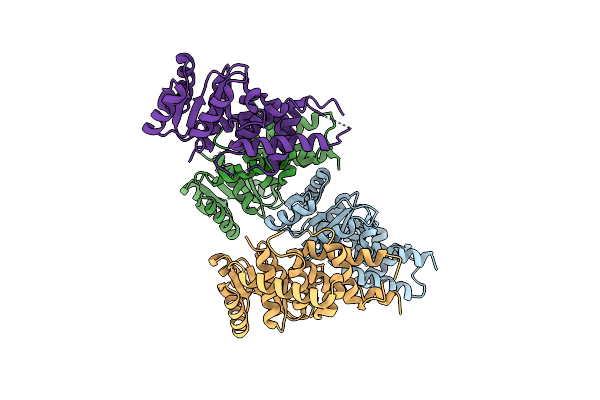 |
Organism: Rubellimicrobium thermophilum dsm 16684
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2025-05-28 Classification: SIGNALING PROTEIN |
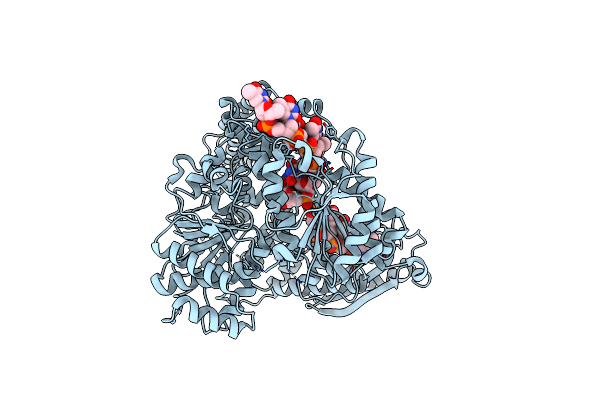 |
Crystal Structure Of Staphylococcus Aureus Ding Protein In Complex With Ssdna And Ca2+
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Chemical production metagenome
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2025-05-21 Classification: HYDROLASE/DNA Ligands: CA |
 |
Organism: Rubellimicrobium thermophilum dsm 16684
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-05-21 Classification: SIGNALING PROTEIN Ligands: MG, NA, TRS |
 |
Crystal Structure Of Bifunctional Glmu From Staphylococcus Aureus Nctc 8325 Complexed With Acetyl Coa And Citrate
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: ACO, CIT |
 |
Crystal Structure Of Bifunctional Glmu From Staphylococcus Aureus Nctc 8325 Complexed With Utp, Coa And Glc 1-P
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: COA, MG, G1P, UTP |
 |
Organism: Candidatus lokiarchaeia archaeon
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: LIPID BINDING PROTEIN |

