Search Count: 39
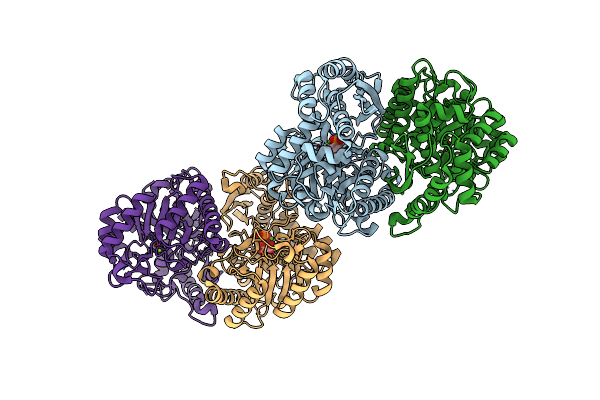 |
Crystal Structure Of Enolase1 From Candida Albicans Complexed With 2'-Phosphoglyceric Acid Sodium
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-06 Classification: METAL BINDING PROTEIN Ligands: MG, 2PG |
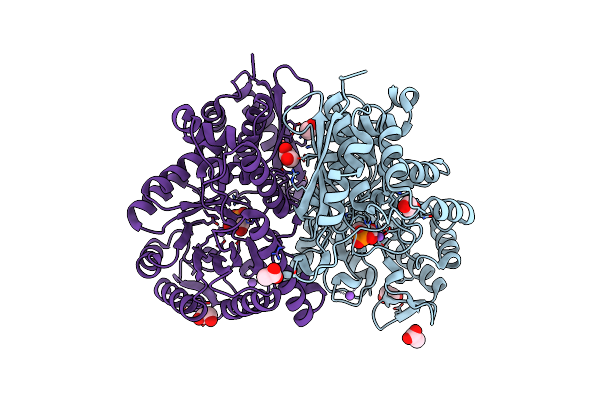 |
Crystal Structure Of Enolase Family Protein From Naegleria Fowleri With Bound 2-Phosphoglyceric Acid
Organism: Naegleria fowleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-04-13 Classification: LYASE Ligands: 2PG, EDO, ACT, NA |
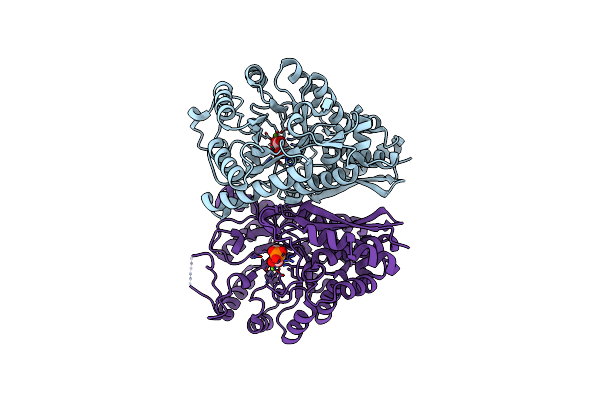 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-03-16 Classification: LYASE Ligands: 2PG, MG |
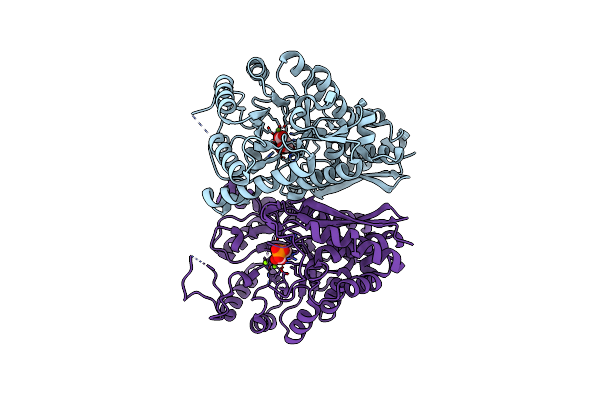 |
Aspergillus Fumigatus Enolase Bound To Phosphoenolpyruvate And 2-Phosphoglycerate
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-16 Classification: LYASE Ligands: MG, 2PG, PEP |
 |
Mycobacterium Tuberculosis Enolase In Complex With Alternate 2-Phosphoglycerate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-01-26 Classification: LYASE Ligands: EDO, ACT, MG, 2PG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-07-28 Classification: LYASE Ligands: MG, 2PG, GOL, ACT, PEG, CL |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: EDO, 2PG, ACT, MG |
 |
Crystal Structure Of Enolase From Legionella Pneumophila Bound To 2-Phosphoglyceric Acid And Magnesium
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-01-23 Classification: LYASE Ligands: 2PG, MG, EDO |
 |
Crystal Structure Of Enolase From Escherichia Coli With Bound 2-Phosphoglycerate Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-10-31 Classification: LYASE Ligands: 2PG, MG, SO4, GOL |
 |
Crystal Structure Of Enolase From E. Coli With A Mixture Of Apo Form, Substrate, And Product Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-10-31 Classification: LYASE Ligands: MG, SO4, GOL, PEP, 2PG, MES |
 |
Thermostable Enolase From Chloroflexus Aurantiacus With Substrate 2-Phosphoglycerate
Organism: Chloroflexus aurantiacus (strain atcc 29366 / dsm 635 / j-10-fl)
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2015-07-01 Classification: LYASE Ligands: MG, 2PG |
 |
Crystal Structure Of Post-Catalytic Binary Complex Of Phosphoglycerate Mutase From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-05-06 Classification: ISOMERASE Ligands: MN, 2PG |
 |
Crystal Structure Of Phosphoglycerate Mutase From Staphylococcus Aureus In 2-Phosphoglyceric Acid Bound Form
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-01-14 Classification: ISOMERASE Ligands: 2PG, MN, DTT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, PEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS, PEP |
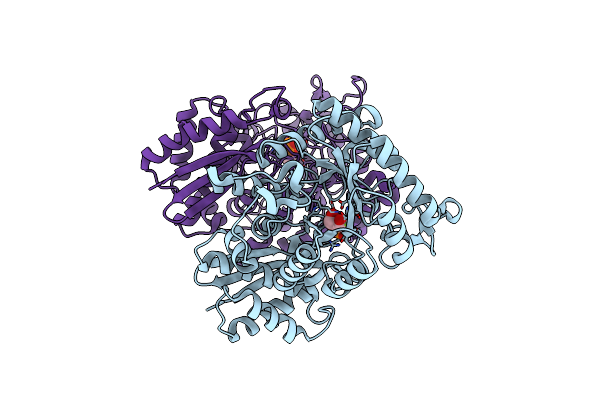 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, PEP |
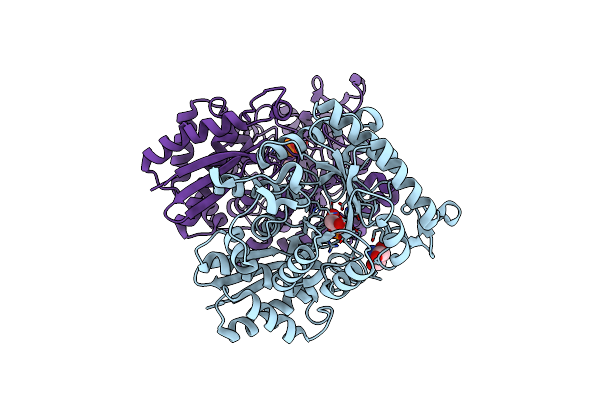 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-08-22 Classification: LYASE Ligands: MG, 2PG, TRS, PEP |
 |
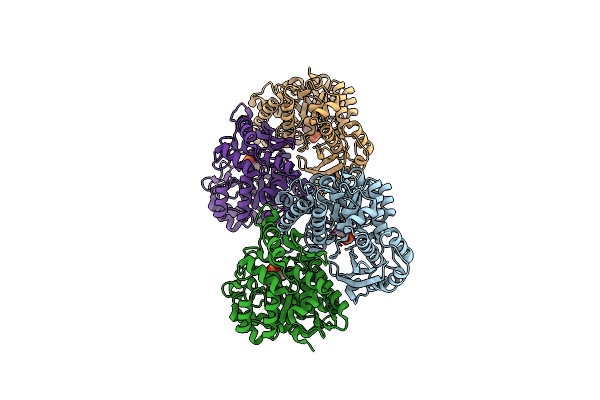
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-13 Classification: LYASE Ligands: 2PG, SO4, MG |
 |
Engineering The Enolase Active Site Pocket: Crystal Structure Of The S39N D321A Mutant Of Yeast Enolase 1
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, 2PG |

