Search Count: 3,348
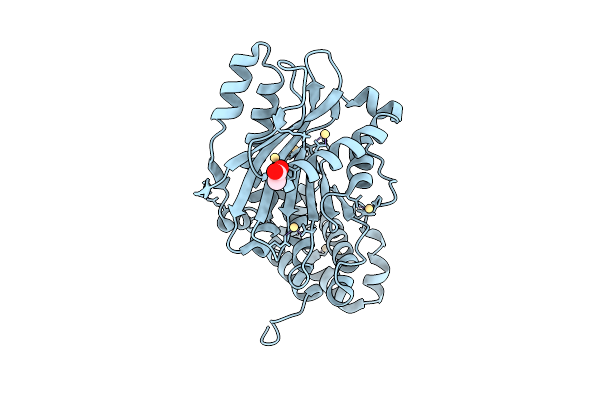 |
Organism: Acinetobacter baylyi adp1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-11-12 Classification: PROTEIN BINDING Ligands: ACT, CD |
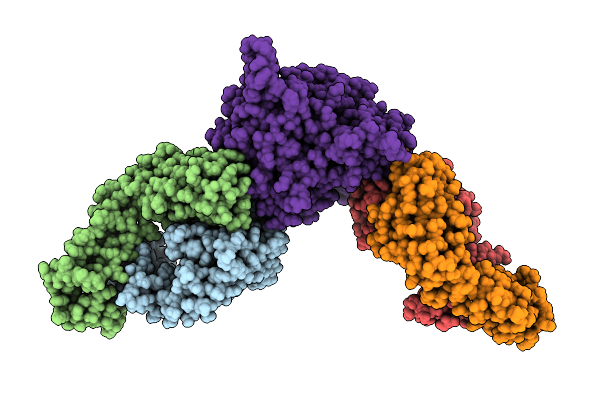 |
Organism: Mus musculus, Langya virus
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
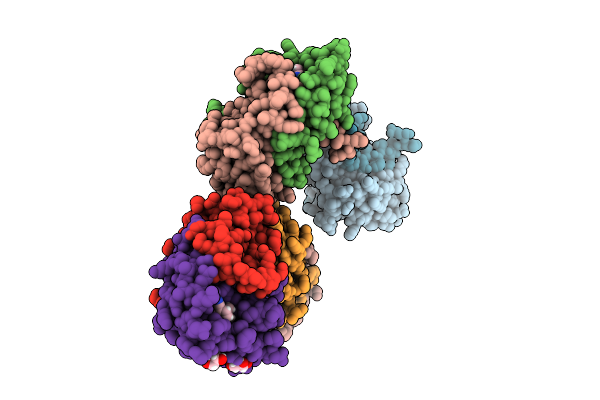 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: A1EAB, PEG, GOL |
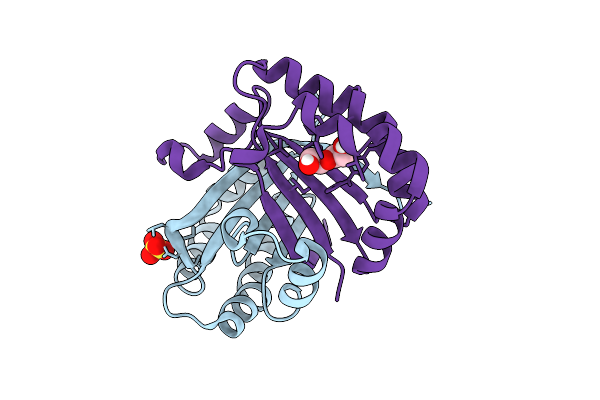 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, PEG |
 |
Organism: Legionella pneumophila str. lens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TOXIN |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL, A1D9Q |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Angavokely Virus (Angv) Fusion (F) Protein Ectodomain In Pre-Fusion Conformation
Organism: Angavokely henipavirus
Method: ELECTRON MICROSCOPY Resolution:4.03 Å Release Date: 2025-03-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Angavokely henipavirus
Method: ELECTRON MICROSCOPY Resolution:5.90 Å Release Date: 2025-03-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Lystt72, A Lytic Endopeptidase From Thermus Thermophilus Mat72 Phage Vb_Tt72
Organism: Thermus phage tt72, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: ZN, PO4 |
 |
Lystt72, A Lytic Endopeptidase From Thermus Thermophilus Mat72 Phage Vb_Tt72
Organism: Thermus phage tt72, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: ZN, PO4 |
 |
Lystt72, A Lytic Endopeptidase From Thermus Thermophilus Mat72 Phage Vb_Tt72
Organism: Thermus phage tt72, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: ZN, PO4 |
 |
Organism: Erythrobacter cryptus dsm 12079
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: MES |
 |
Organism: Mangifera indica
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Mangifera indica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Crystal Structure Of K38 Amylase From Bacillus Sp. Strain Ksm-K38 Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacillus sp. ksm-k38
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, ACT, NA |

