Search Count: 46
 |
The X-Ray Crystallographic Structure Of Branching Enzyme From Rhodothermus Obamensis Stb05
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-03-04 Classification: CARBOHYDRATE |
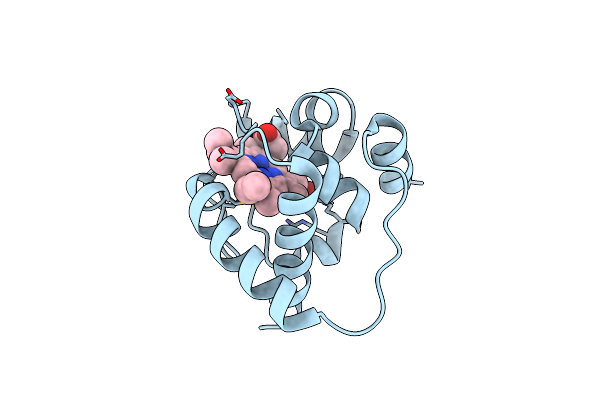 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2018-06-27 Classification: LYASE Ligands: HEC |
 |
Engineered Cytochrome C From Rhodothermus Marinus (Rma Tde) Bound To Carbene Intermediate (1-Ethoxy-1-Oxopropan-2-Ylidene)
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2018-06-27 Classification: LYASE Ligands: HEC, CA1 |
 |
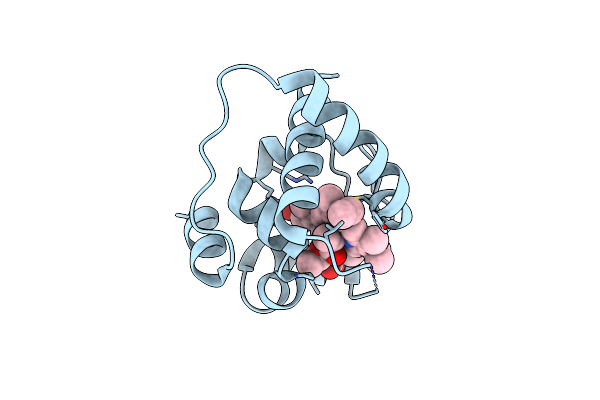
Crystal Structure Of The Esterase Domain From Rhodothermus Marinus Rmar_1206 Protein
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: PGE |
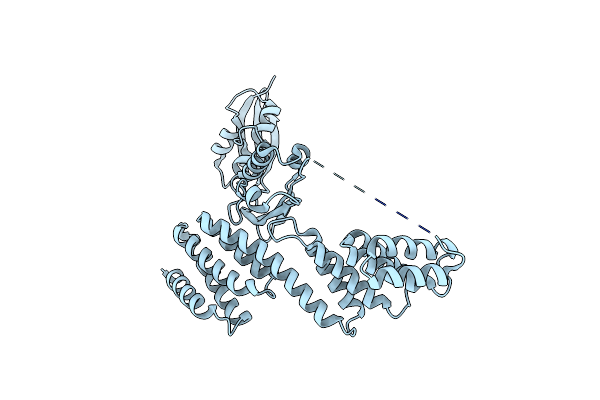 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-27 Classification: CELL ADHESION |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.6 Å, 1.606 Å Release Date: 2015-10-28 Classification: SUGAR BINDING PROTEIN Ligands: CA, DOD |
 |
Xyloglucan Binding Module (Cbm4-2 X2-L110F) In Complex With Branched Xyloses
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: CA, GLC |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2013-12-25 Classification: ISOMERASE Ligands: PO4, CL |
 |
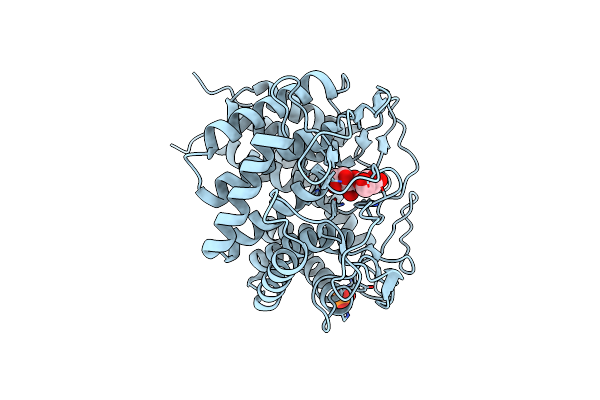
Crystal Structure Of Cellobiose 2-Epimerase In Complex With Glucosylmannose
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2013-12-25 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-12-25 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2013-12-25 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Xylopentaose Binding Mutated (X-2 L110F) Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA |
 |
Cellopentaose Binding Mutated (X-2 L110F) Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA |
 |
X-2 L110F Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA |
 |
X-2 Engineered Mutated Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA |
 |
Xylotetraose Bound To X-2 Engineered Mutated Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA, CIT |
 |
Xylopentaose Binding X-2 Engineered Mutated Cbm4-2 Carbohydrate Binding Module From A Thermostable Rhodothermus Marinus Xylanase
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2012-03-07 Classification: HYDROLASE Ligands: CA |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-02 Classification: TRANSFERASE Ligands: LAC, TRS, NA |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-08-18 Classification: HYDROLASE Ligands: GOL, CA |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2009-12-15 Classification: ELECTRON TRANSPORT Ligands: SF4 |

