Search Count: 661
 |
Crystal Structure Of Rid Family Protein Aciad3089 From Acinetobacter Baylyi
Organism: Acinetobacter baylyi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Crystal Structure Of Rid Family Protein Aciad3089 From Acinetobacter Baylyi In C2 Space Group
Organism: Acinetobacter baylyi
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
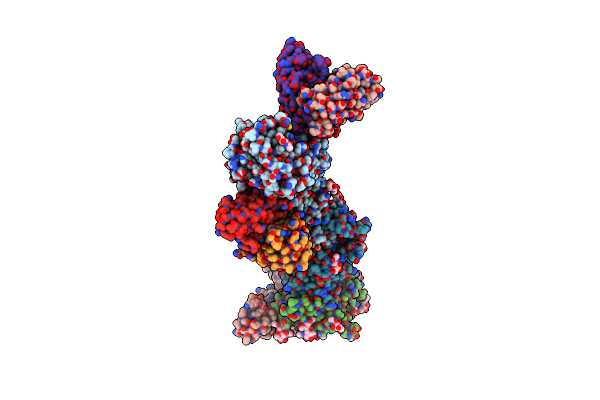
Cryoem Structure Of The Hcmv Trimer Ghglgo In Complex With Neutralizing Fabs 13H11 And Msl-109
Organism: Human cytomegalovirus (strain merlin), Human cytomegalovirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: VIRAL PROTEIN/Immune System Ligands: NAG, MAN |
 |
Cryoem Structure Of The Hcmv Trimer Ghglgo In Complex With Human Platelet-Derived Growth Factor Receptor Alpha And Neutralizing Fabs 13H11 And Msl-109
Organism: Human cytomegalovirus (strain merlin), Human cytomegalovirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
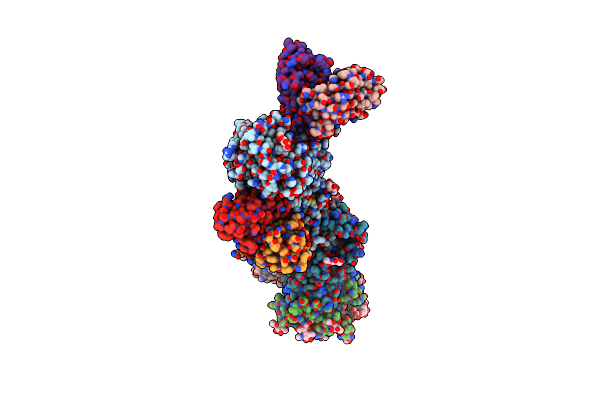 |
Cryoem Structure Of The Hcmv Trimer Ghglgo In Complex With Human Transforming Growth Factor Beta Receptor Type 3 And Neutralizing Fabs 13H11 And Msl-109
Organism: Human cytomegalovirus (strain merlin), Human cytomegalovirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
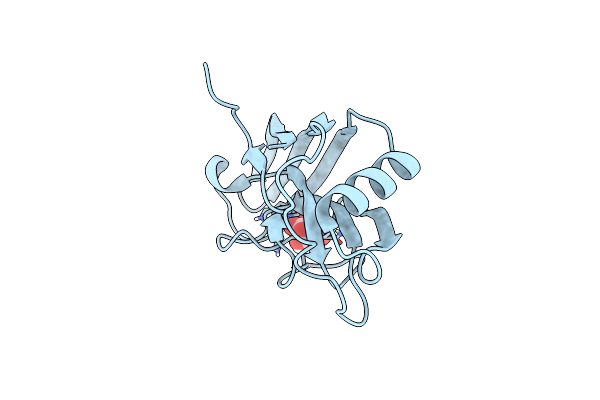 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, TLA |
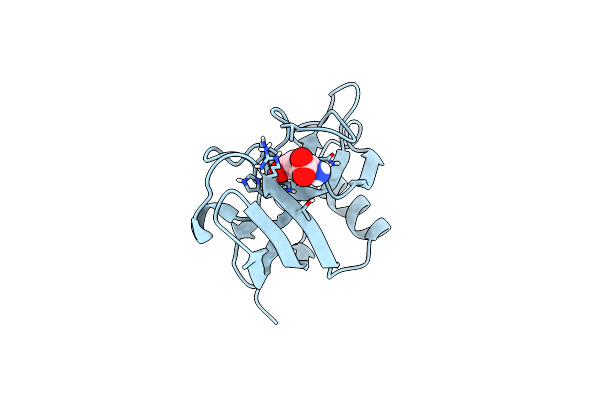 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, GLU |
 |
Crystal Structure Of The Acyl-Carrier-Protein Udp-N-Acetylglucosamine O-Acyltransferase Lpxa From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: SO4, PO3 |
 |
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-26 Classification: LYASE Ligands: SO4 |
 |
Organism: Proteus mirabilis hi4320
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: GOL, PO4 |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-211
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: CO |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-217
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: SO4 |
 |
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |
 |
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The N-Terminal Periplasmic Domain Of Scsb From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Hcmv Pentamer In Complex With Neutralizing Antibody 8I21
Organism: Human cytomegalovirus, Human cytomegalovirus (strain 5508), Human cytomegalovirus (strain ad169), Human cytomegalovirus (strain merlin), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2017-07-05 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Crystal Structure Of Hcmv Pentamer In Complex With Neutralizing Antibody 8I21 - Low Resolution Dataset For Initial Phasing By Sad
Organism: Human cytomegalovirus, Human cytomegalovirus (strain 5508), Human cytomegalovirus (strain ad169), Human cytomegalovirus (strain merlin), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.99 Å Release Date: 2017-07-05 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Crystal Structure Of Hcmv Pentamer In Complex With Neutralizing Antibody 9I6
Organism: Human cytomegalovirus, Human cytomegalovirus (strain 5508), Human cytomegalovirus (strain ad169), Human cytomegalovirus (strain merlin), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:5.90 Å Release Date: 2017-07-05 Classification: Viral Protein/Immune System |
 |
Crystal Structure Of Universal Stress Protein E From Proteus Mirabilis In Complex With Udp-3-O-[(3R)-3-Hydroxytetradecanoyl]-N-Acetyl-Alpha-Glucosamine
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-26 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: U20, GOL, CL, SO4 |
 |
Crystal Structure Of Proteus Mirabilis Transcriptional Regulator Protein Crl At 1.95A Resolution
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-08-06 Classification: Transcription regulator Ligands: NHE |

