Search Count: 118
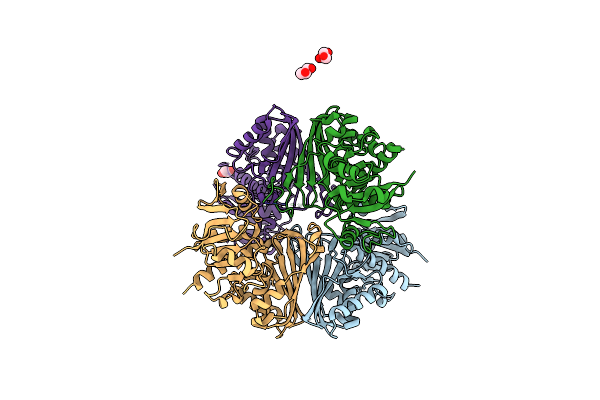 |
Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Candida Albicans
Organism: Candida albicans wo-1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: GOL |
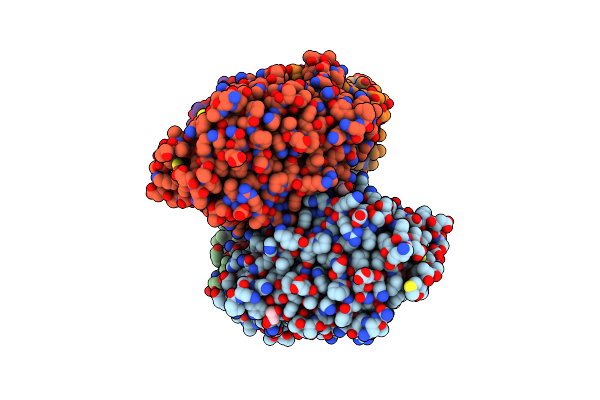 |
Crystal Structure Of Candida Albicans Calcineurin A, Calcineurin B, Fkbp12 And Fk506 (Tacrolimus)
Organism: Candida albicans (strain wo-1), Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-09-18 Classification: Hydrolase/Isomerase/Calcium Binding Ligands: PO4, ZN, FE, EDO, CA, FK5 |
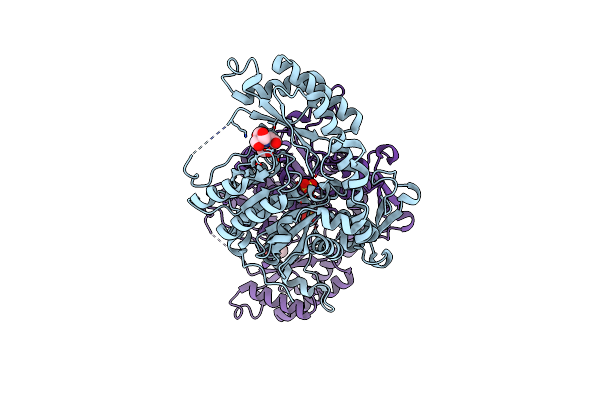 |
Organism: Candida albicans wo-1
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2019-02-20 Classification: TRANSFERASE Ligands: BTB, SO4, CL |
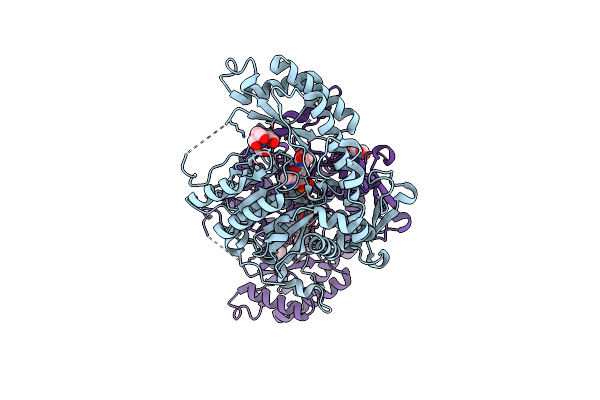 |
Crystal Structure Of The Aminotransferase Aro8 From C. Albicans With Ligands
Organism: Candida albicans wo-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-02-20 Classification: TRANSFERASE Ligands: BTB, PLP, PHE, CL, TRS |
 |
Organism: Candida albicans (strain wo-1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-19 Classification: TRANSFERASE Ligands: MES, COA |
 |
Organism: Candida albicans wo-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2017-04-19 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: COA |
 |
Structure Of Candida Albicans Kar3 Motor Domain Fused To Maltose-Binding Protein
Organism: Escherichia coli, Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-10-10 Classification: MOTOR PROTEIN Ligands: MG, ADP, EDO |
 |
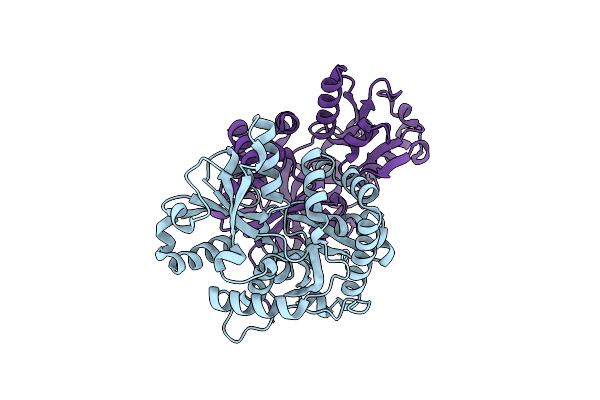
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-09-19 Classification: TRANSFERASE Ligands: CIT |
 |
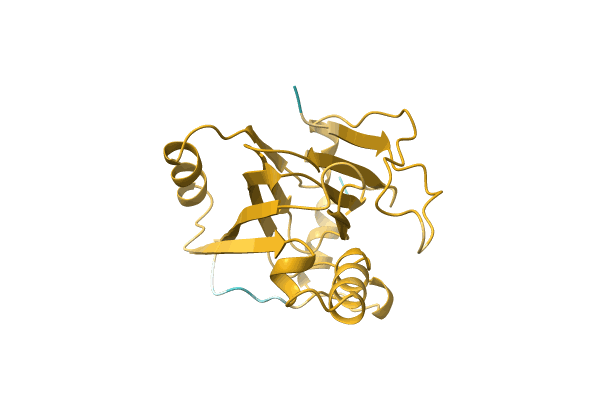
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-09-19 Classification: TRANSFERASE |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
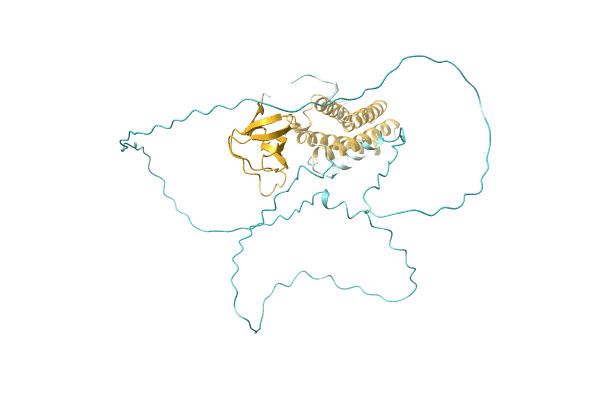
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
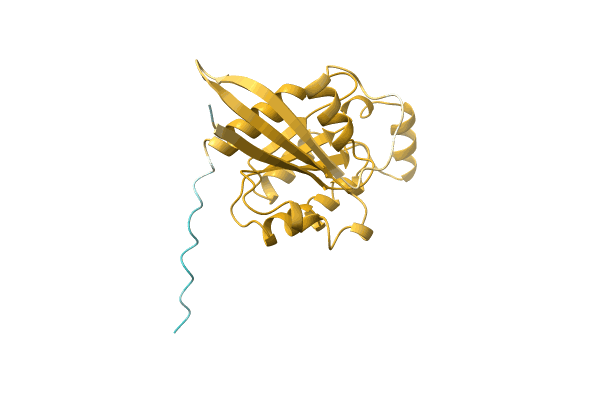
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
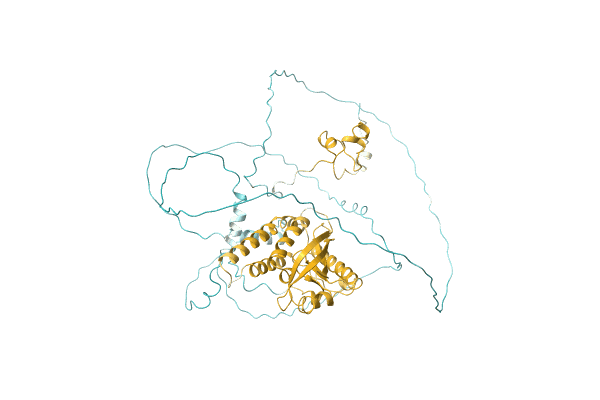
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
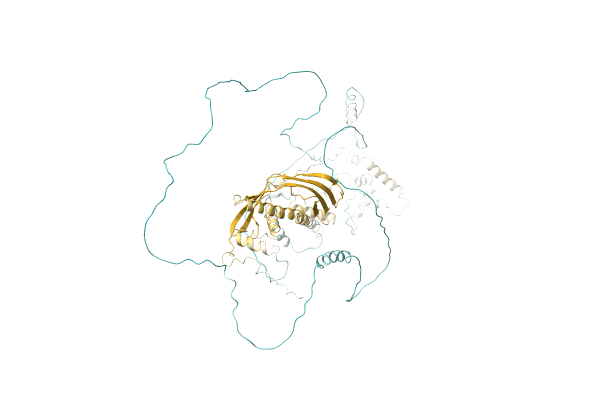
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Candida albicans (strain WO-1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

