Search Count: 4,812
 |
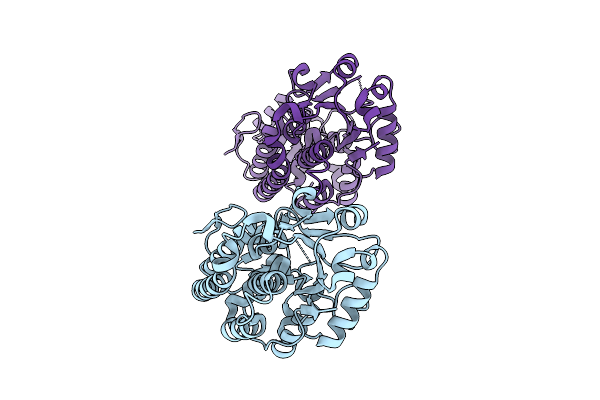
Structure And Mechanism Of The Broad Spectrum Crispr-Associated Ring Nuclease Crn4
Organism: Hyphomicrobiales bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN |
 |
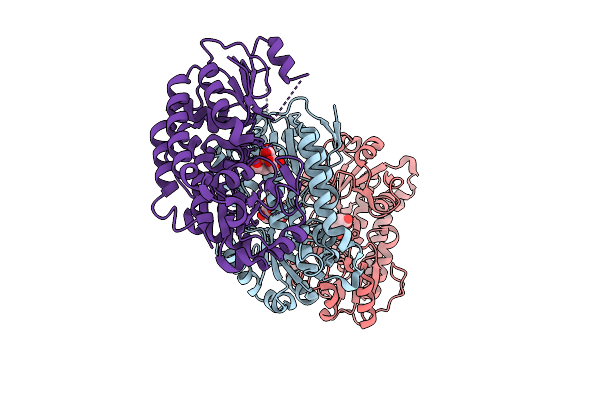
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN |
 |
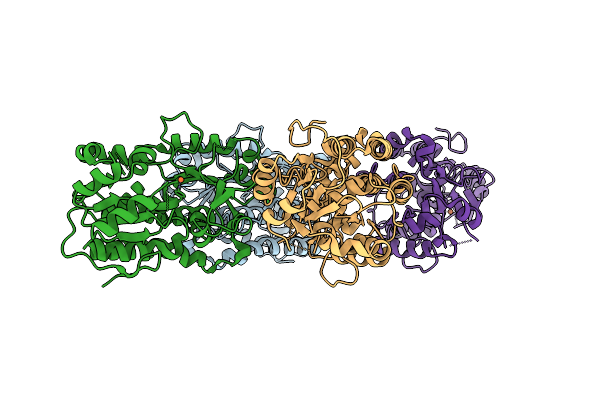
Structure Of Yiua From Yersinia Ruckeri With Iron And Nitrilotriacetic Acid
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: NTA, FE |
 |
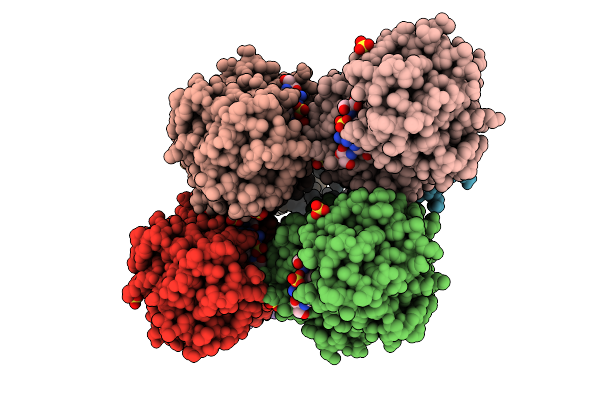
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE, A1I15, SO4 |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: EDO, MES |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: MG, CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: CL |
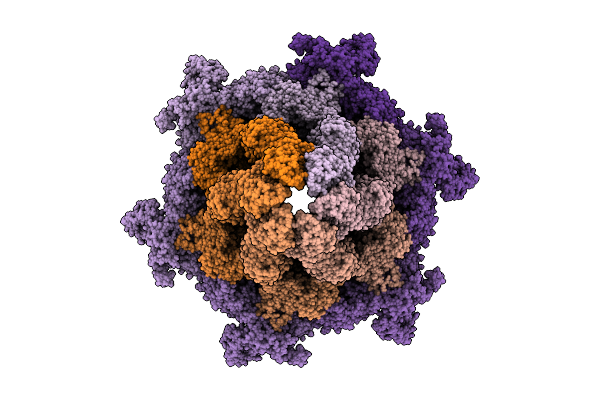 |
Cryo-Em Structure Of The Plpvc1 Baseplate, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
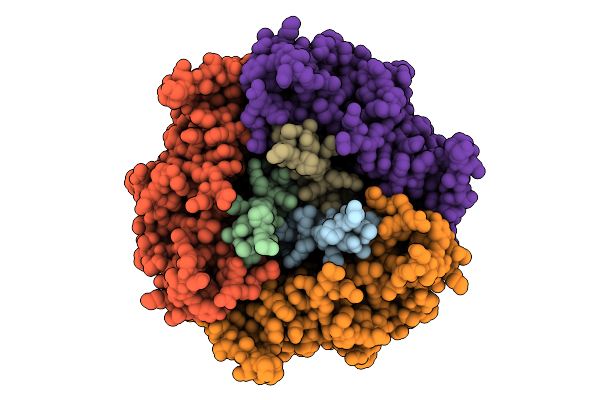 |
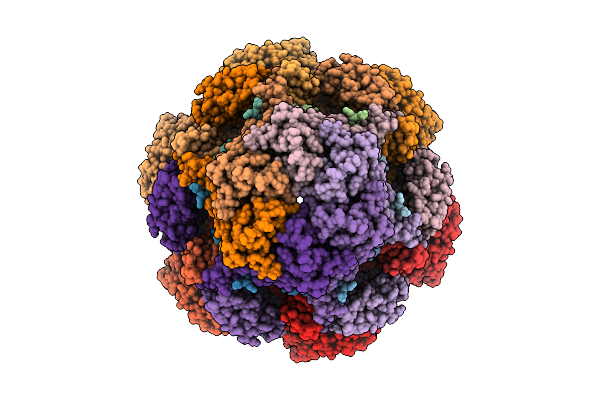
Cryo-Em Structure Of The Plpvc1 Central Spike, 3-Fold Symmetrized (C3), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
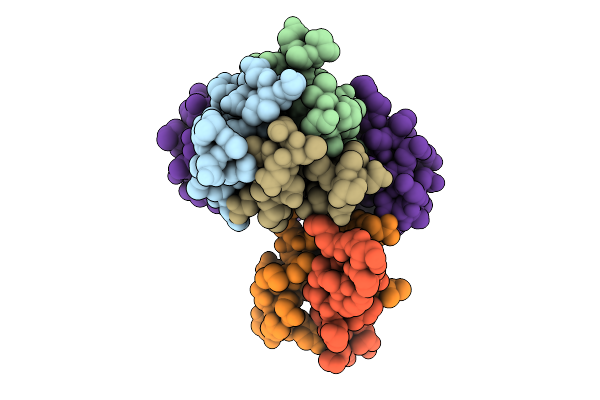
Cryo-Em Structure Of The Plpvc1 Cap, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
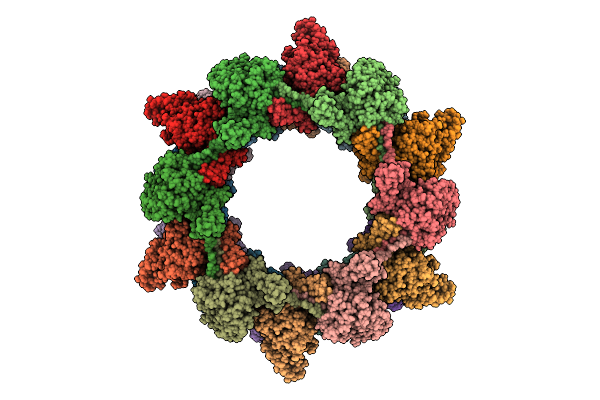
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
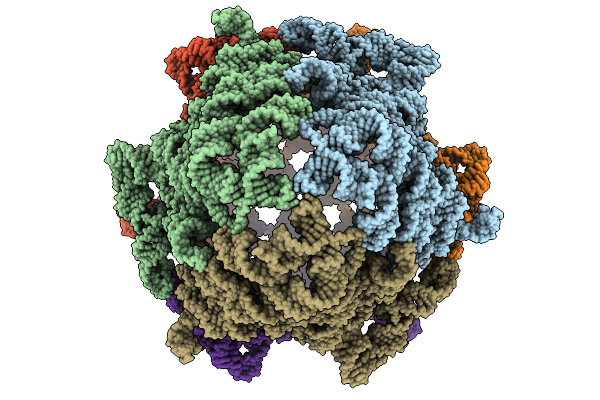
Cryo-Em Structure Of The Plpvc1 Sheath, 6-Fold Symmetrized (C6), In Contracted State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Brucella melitensis
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN |
 |
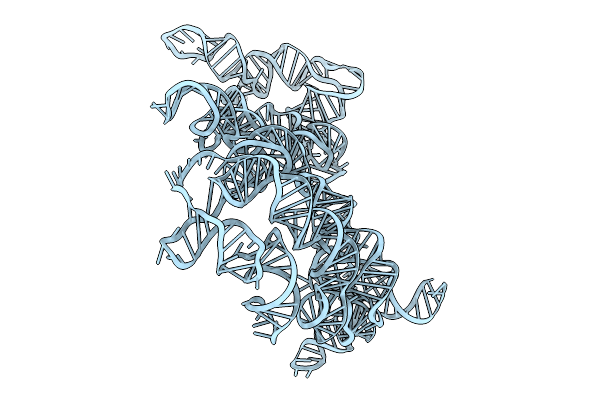
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
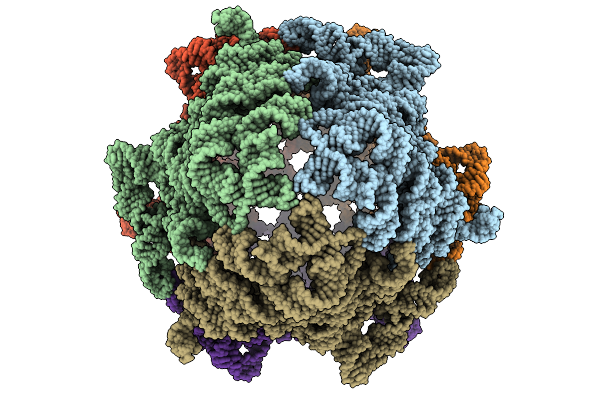
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
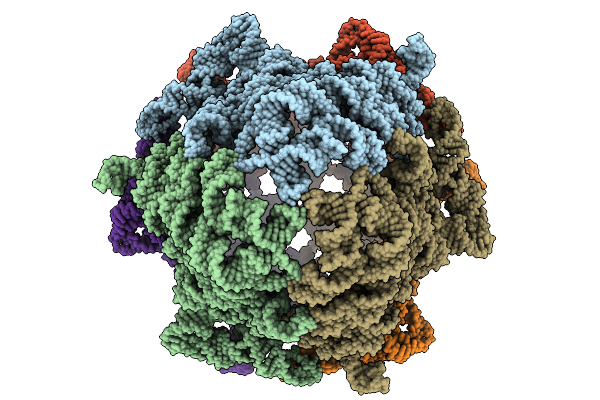
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
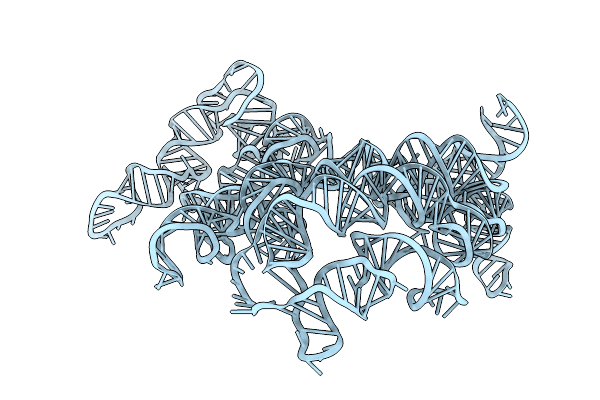
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Kynurenine Monooxygenase From Pseudomonas Fluorescens Complexed With 4-Cyclohexylphenylacetyltetrazole
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: FLAVOPROTEIN/INHIBITOR Ligands: A1AIN, FAD, CL, DMS |

