Search Count: 11,684
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: HYDROLASE |
 |
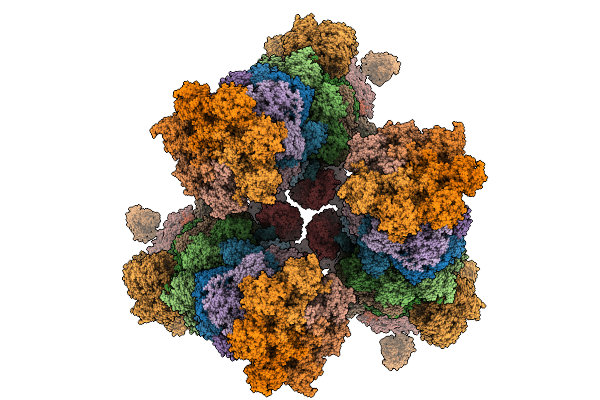
Pentamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate State2
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
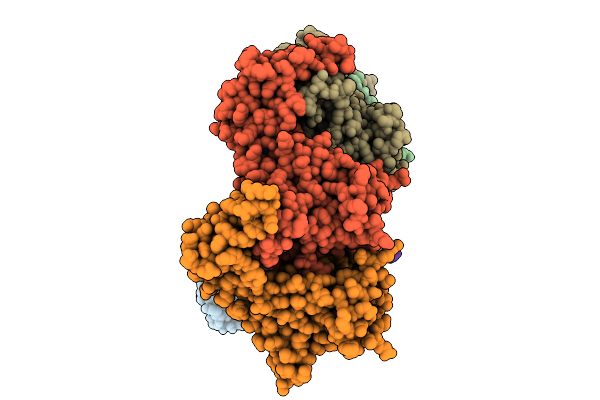
Sixteen Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
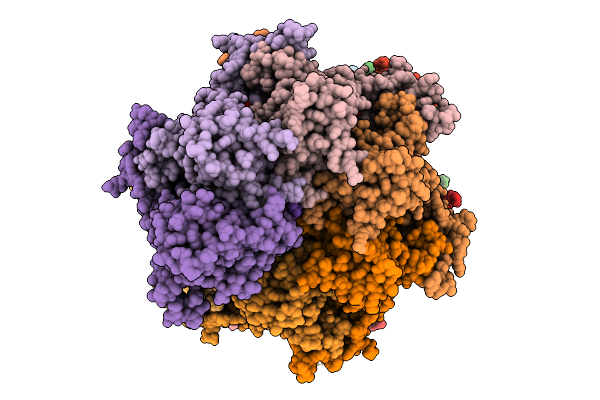 |
Twenty-Two Polymer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
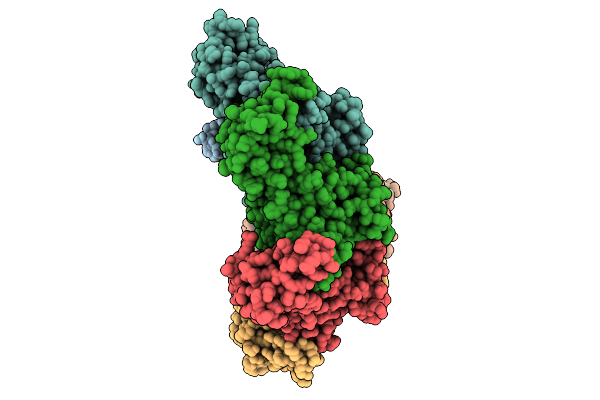 |
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
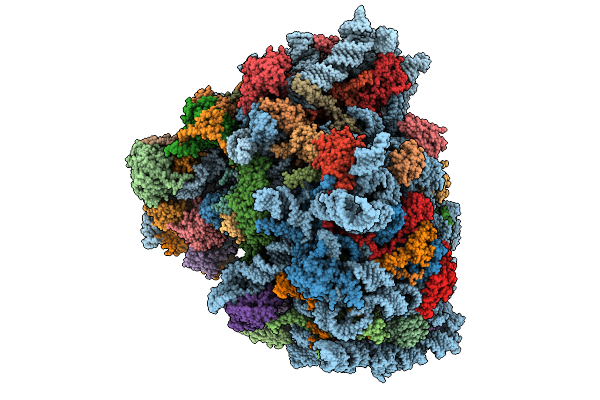 |
Rabbit 80S Ribosome In Complex With Erf1-Aaq, Stalled At The Stop Codon In Mutated F2A Sequence
Organism: Homo sapiens, Foot-and-mouth disease virus sat 2, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RIBOSOME Ligands: MG, ZN, K, SPD, GTP, SF4 |
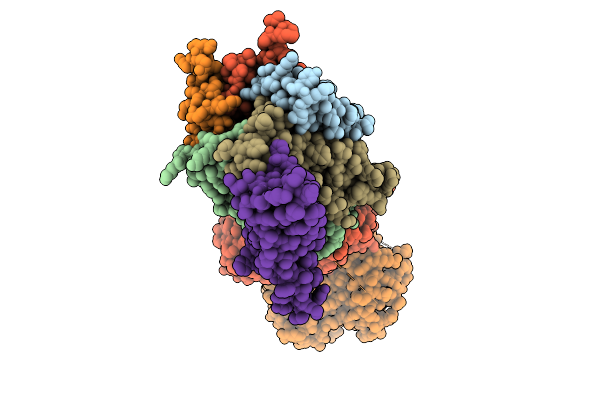 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2026-01-21 Classification: TRANSPORT PROTEIN Ligands: IHP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-01-14 Classification: SPLICING |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-14 Classification: SPLICING Ligands: HOH |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-14 Classification: SPLICING |
 |
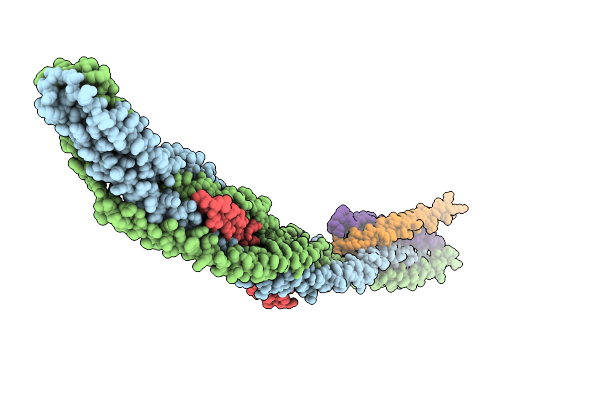
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Resolution:2.87 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2026-01-14 Classification: GENE REGULATION |
 |
Organism: Lyssavirus rabies, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
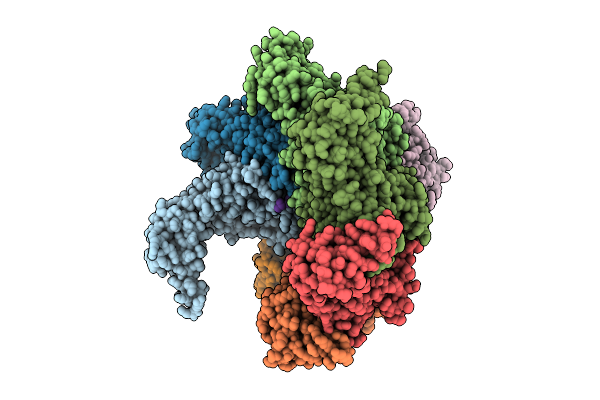
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
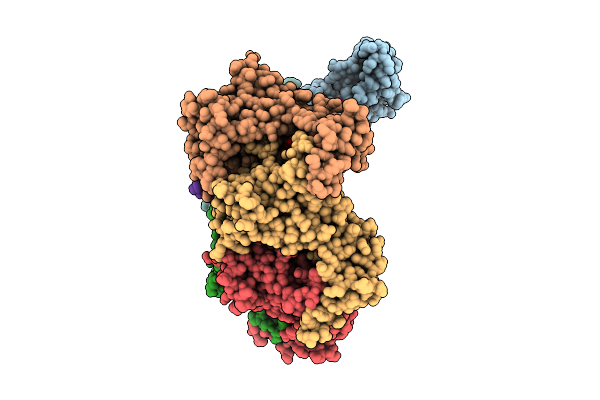
Octamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:3.17 Å Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
 |
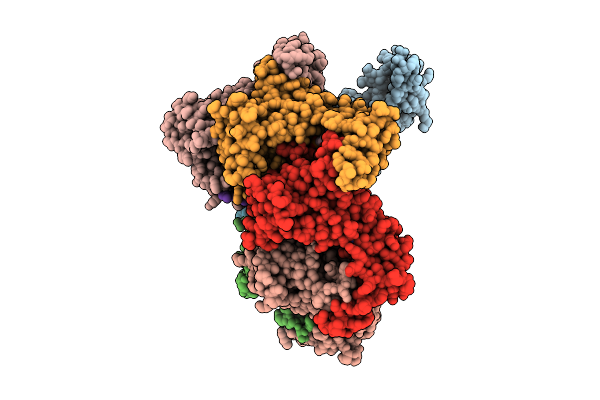
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Escherichia coli bl21(de3), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Heptamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
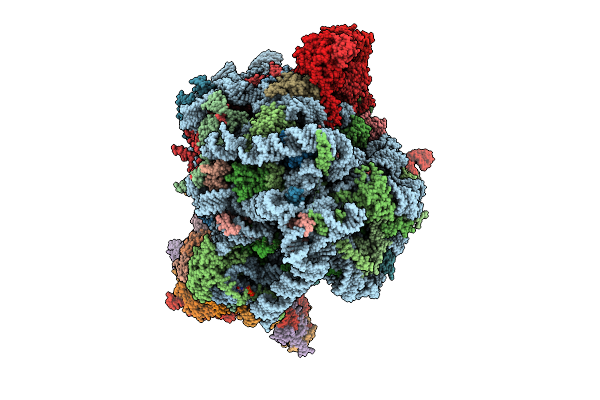 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
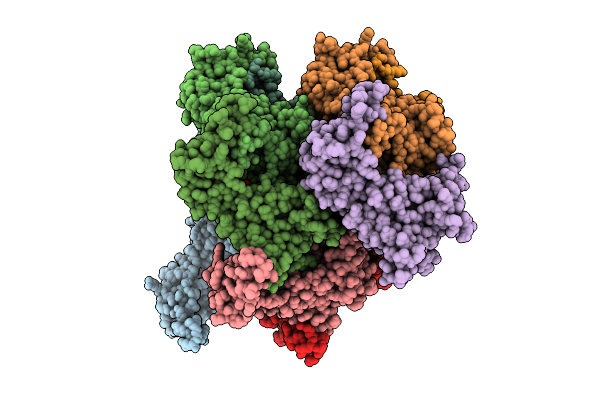 |
Eleven Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutaion) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:2.98 Å Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |

