Search Count: 336
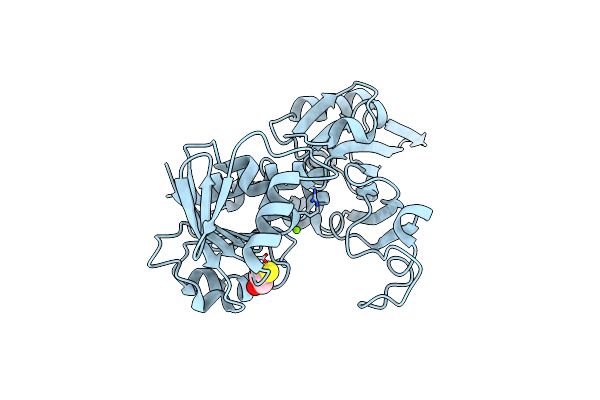 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase Gapdh From Clostridium Beijerinckii
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-03-17 Classification: OXIDOREDUCTASE Ligands: BME, MG |
 |
Crystal Structure Of A Beta-1,3-Glucanase Domain (Gh64) From Clostridium Beijerinckii
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-08 Classification: HYDROLASE |
 |
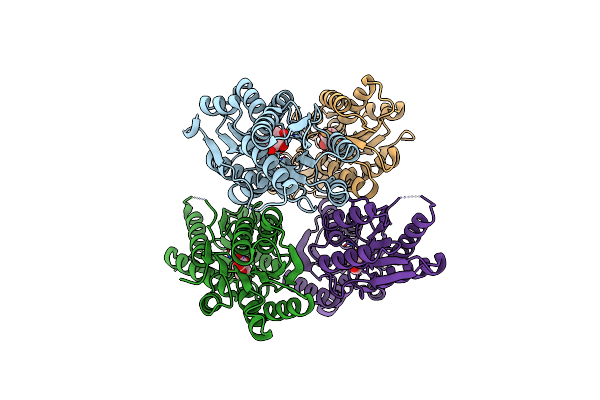
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN Ligands: XYP |
 |
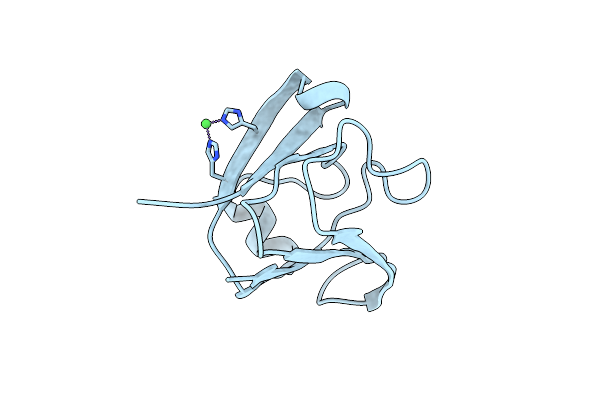
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN Ligands: XYP |
 |
Crystal Structure Of Clostrillin Double Mutant (S17H,S19H) In Complex With Nickel
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-02-01 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
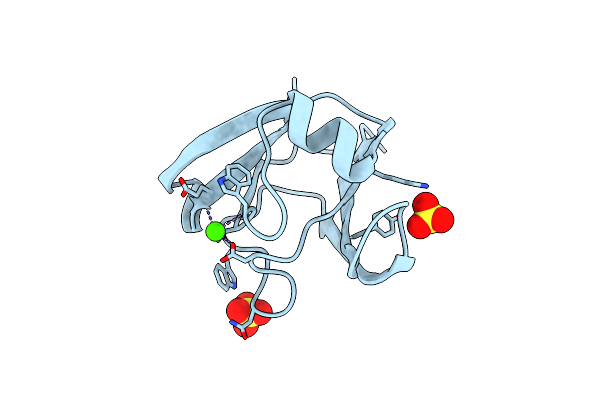
Crystal Structure Of Enolase Superfamily Member From Clostridium Beijerincki Complexed With Mg
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-05-23 Classification: ISOMERASE Ligands: MG |
 |
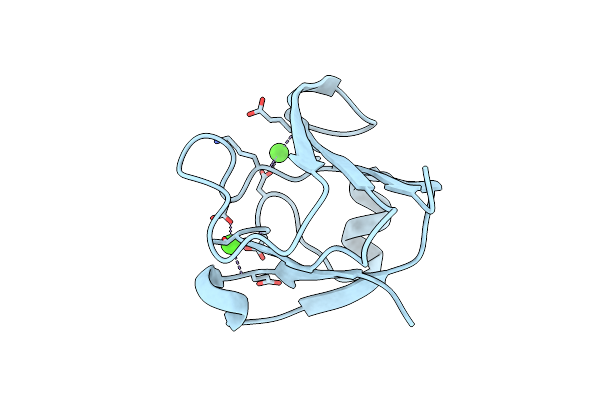
Crystal Structure Of A Mutant T82R Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA, SO4 |
 |
Crystal Structure Of A Mutant W39D Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
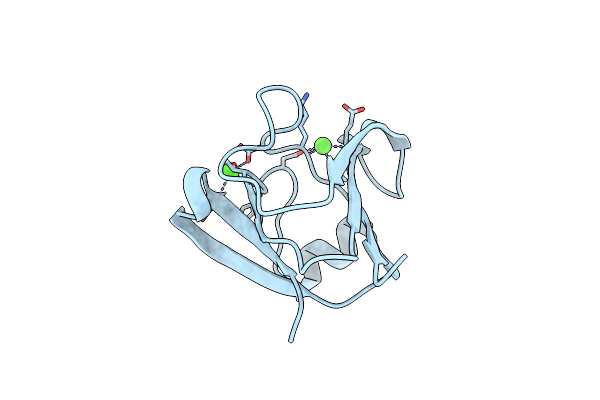
Crystal Structure Of A Mutant T41S Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of A Double Mutant T41S T82S Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA, SO4 |
 |
Crystal Structure Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-12-01 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii-In Alternate Space Group I422
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-01 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Putative Mandelate Racemase/Muconate Lactonizing Protein From Clostridium Beijerinckii Ncimb 8052
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-04-14 Classification: ISOMERASE Ligands: MG |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

