Search Count: 685
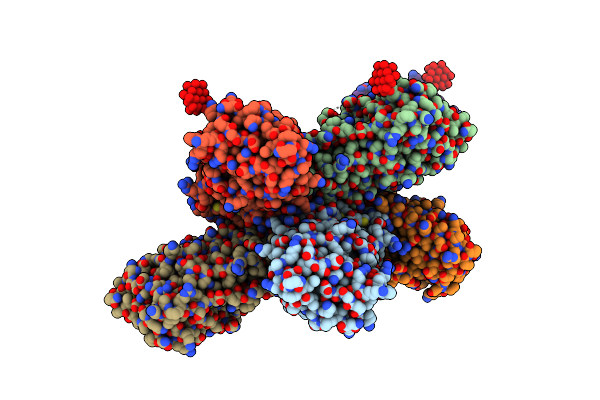 |
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2022-05-18 Classification: METAL BINDING PROTEIN Ligands: ZN, TEW |
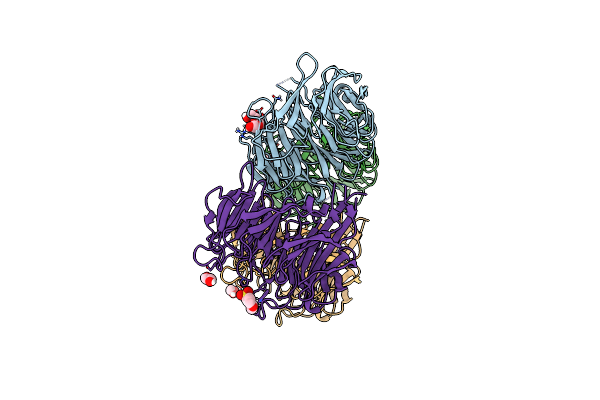 |
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-09-18 Classification: CHAPERONE Ligands: PGE, EDO, MES |
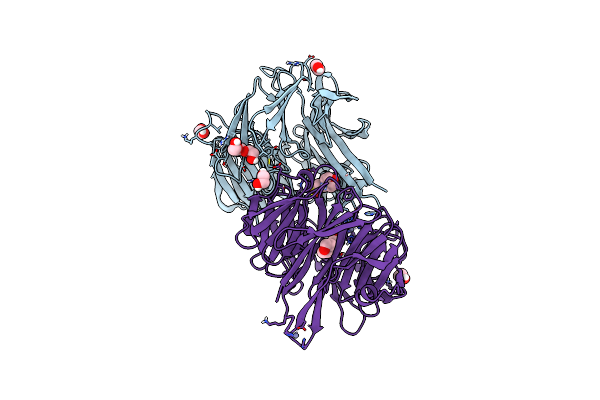 |
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-03-13 Classification: METAL BINDING PROTEIN Ligands: MES, ZN, EDO, PGE, PEG |
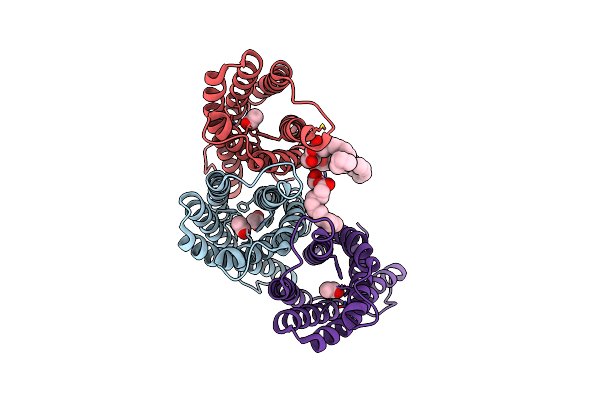 |
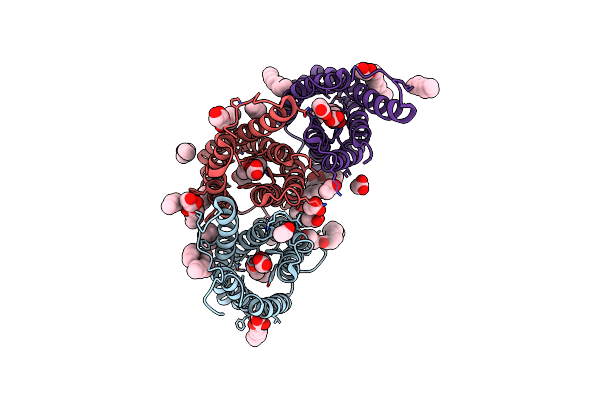
2.8 Angstrom Crystal Structure Of Succinate-Acetate Permease From Citrobacter Koseri
Organism: Citrobacter koseri atcc baa-895
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: ACT, 78M |
 |
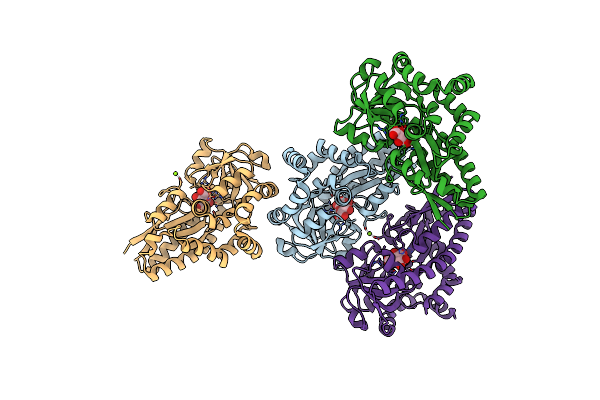
1.8 Angstrom Crystal Structure Of Succinate-Acetate Permease From Citrobacter Koseri
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-04-25 Classification: TRANSPORT PROTEIN Ligands: ACT, PLM, 78M |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Citrobacter Koseri (Cko_04899, Target Efi-510094) With Bound D-Glucuronate
Organism: Citrobacter koseri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-12-10 Classification: SOLUTE BINDING PROTEIN Ligands: BDP, CL, MG |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Citrobacter Koseri (Cko_04899), Target Efi-510094, Apo, Open Structure
Organism: Citrobacter koseri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-11-20 Classification: TRANSPORT PROTEIN |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
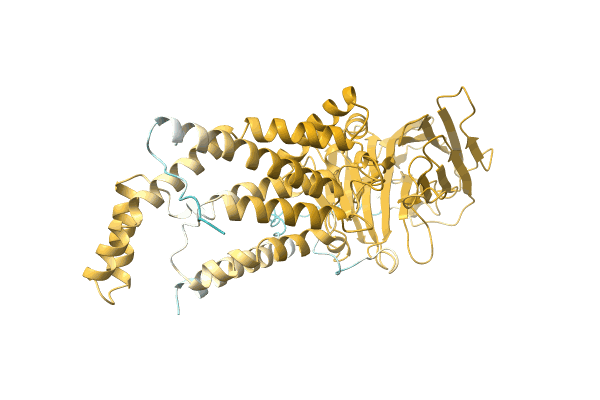 |
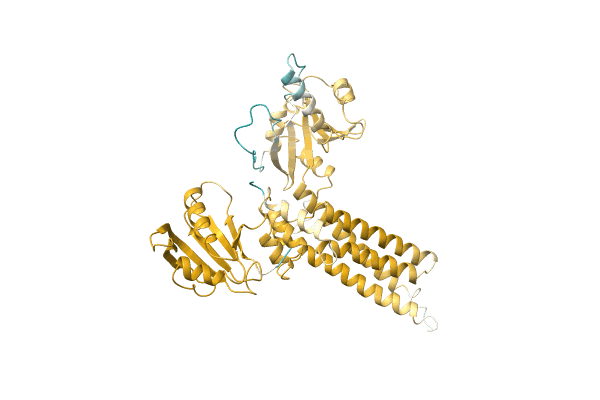
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
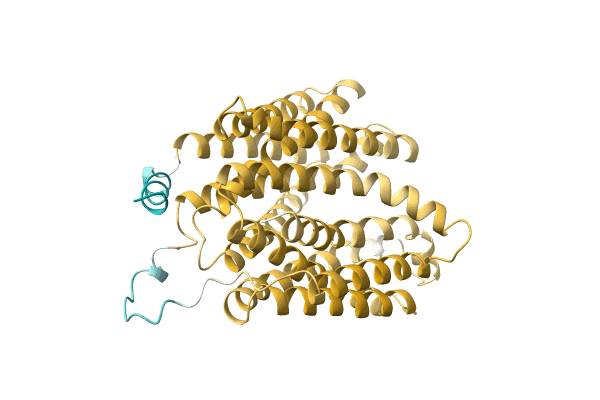
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
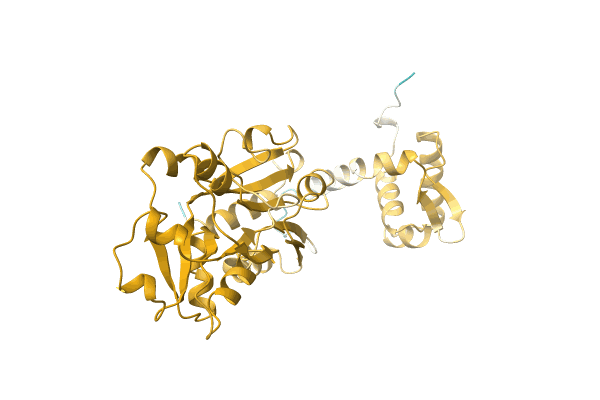
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

