Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
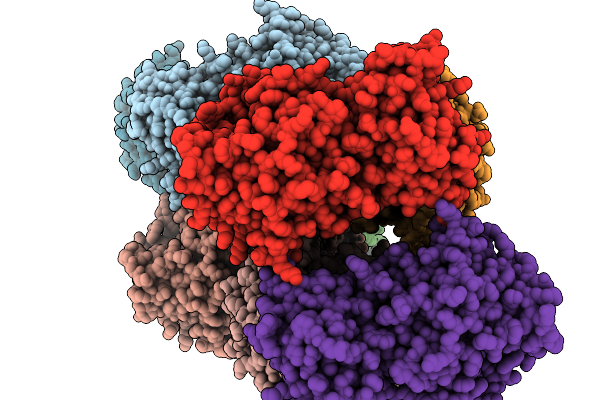 |
Crystal Structure Of Pseudopedobacter Saltans Gh43 Beta-Xylosidase In Complex With Xylose.
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE Ligands: CA, XLS |
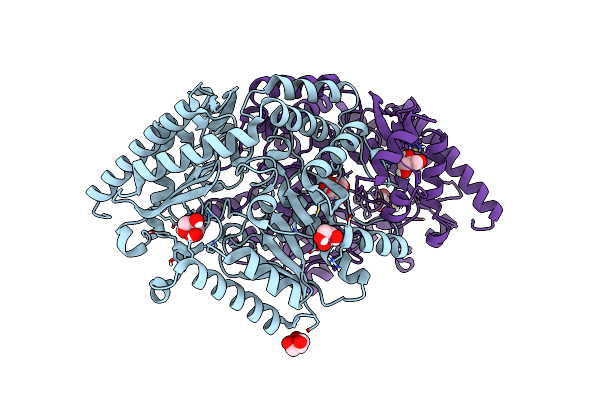 |
Crystal Structure Of The Dimeric Transaminase Doed From C. Salexigens Dsm 3043.
Organism: Chromohalobacter israelensis dsm 3043
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: GOL, PLP |
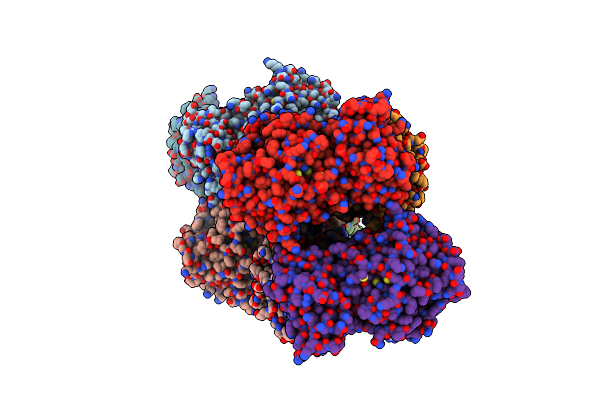 |
Structure Of Gh43 Family Enzyme, Xylan 1, 4 Beta- Xylosidase From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: CA |
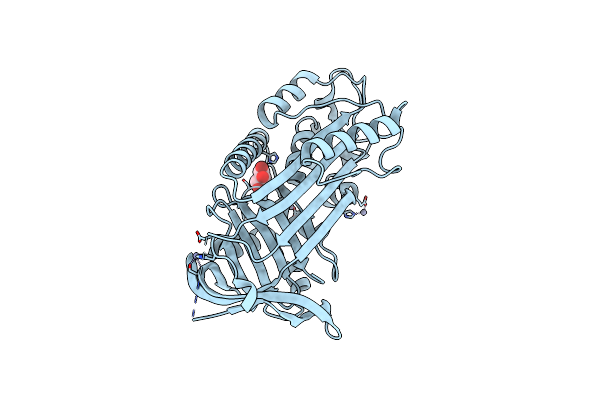 |
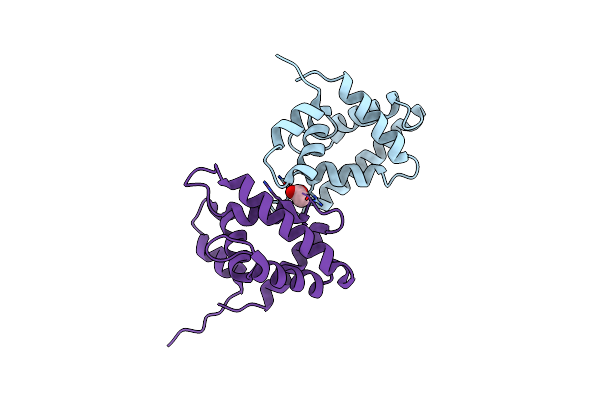
Structure Of A Bacterial Serpin Choloropin Derived From Cholorobium Limicola
Organism: Chlorobium limicola (strain dsm 245 / nbrc 103803 / 6330)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: ZN, GOL |
 |
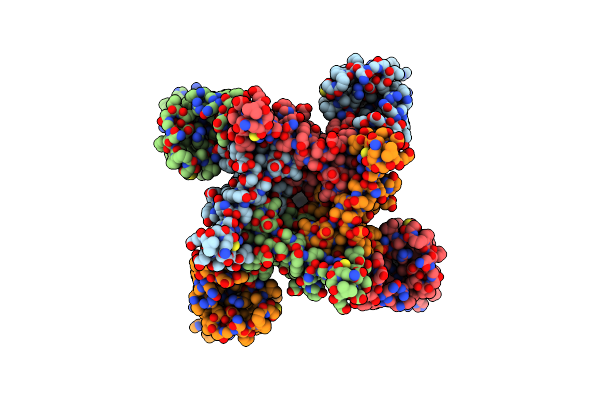
The Crystal Structure Of 15Kda Phlebotomus Papatasi Salivary Protein Ppsp15.
Organism: Phlebotomus papatasi
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2023-04-05 Classification: PROTEIN BINDING Ligands: ACT |
 |
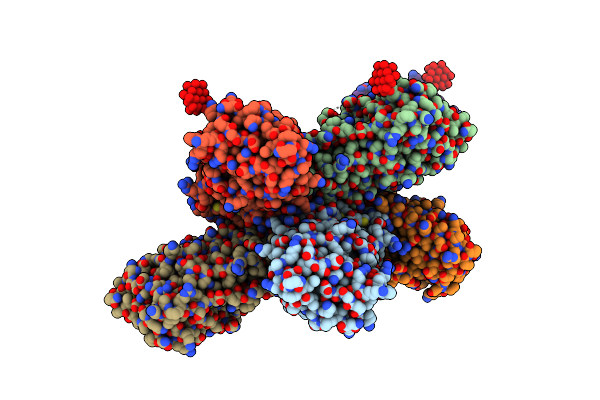
Organism: Emiliania huxleyi
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: TRANSPORT PROTEIN |
 |
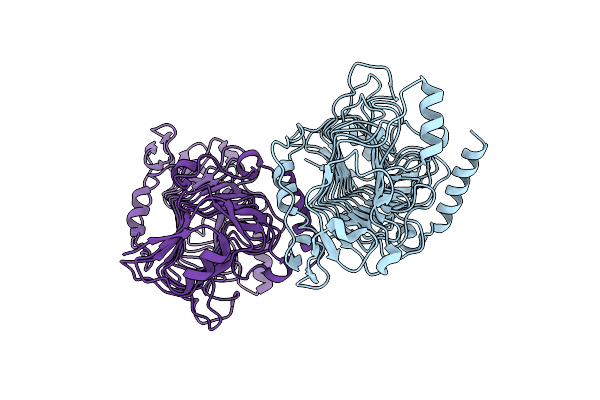
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2022-05-18 Classification: METAL BINDING PROTEIN Ligands: ZN, TEW |
 |
Organism: Chromohalobacter salexigens (strain atcc baa-138 / dsm 3043 / cip 106854 / ncimb 13768 / 1h11)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-10-06 Classification: TRANSPORT PROTEIN Ligands: MG, DHC |
 |
Organism: Chromohalobacter salexigens (strain atcc baa-138 / dsm 3043 / cip 106854 / ncimb 13768 / 1h11)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-10-06 Classification: TRANSPORT PROTEIN Ligands: EDO, HC4, MG |
 |
Organism: Chromohalobacter salexigens (strain atcc baa-138 / dsm 3043 / cip 106854 / ncimb 13768 / 1h11)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-06 Classification: TRANSPORT PROTEIN Ligands: FER, MG, SO4 |
 |
Organism: Chromohalobacter salexigens (strain atcc baa-138 / dsm 3043 / cip 106854 / ncimb 13768 / 1h11)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-10-06 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Structure Of The Pl6 Family Chondroitinase B From Pseudopedobacter Saltans, Pedsa3807
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-07-28 Classification: LYASE |
 |
Structure Of The Pl6 Family Polysaccharide Lyase Pedsa3628 From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-07-28 Classification: LYASE Ligands: PO4 |
 |
Structure Of The Pl6 Family Alginate Lyase Pedsa0632 From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-07-28 Classification: LYASE |
 |
Structure Of The Pl6 Family Alginate Lyase Pedsa0632 From Pseudopedobacter Saltans In Complex With Substrate
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-07-28 Classification: LYASE |
 |
The First Crystal Structure Of The Daba Aminotransferase Ectb In The Ectoine Biosynthesis Pathway Of The Model Organism Chromohalobacter Salexigens Dsm 3034
Organism: Chromohalobacter salexigens (strain dsm 3043 / atcc baa-138 / ncimb 13768)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-03-11 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-09-18 Classification: CHAPERONE Ligands: PGE, EDO, MES |
 |
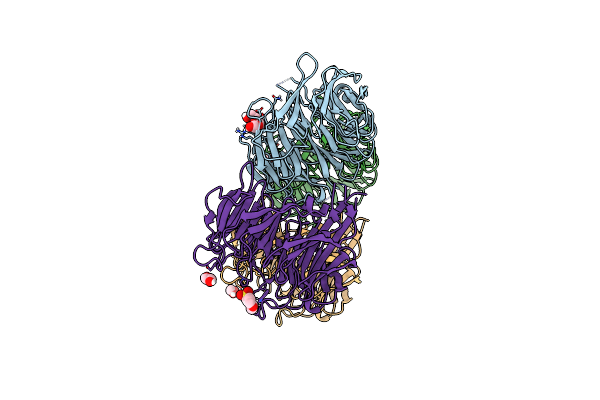
Organism: Porcine epidemic diarrhea virus (strain cv777)
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: VIRAL PROTEIN Ligands: NAG |
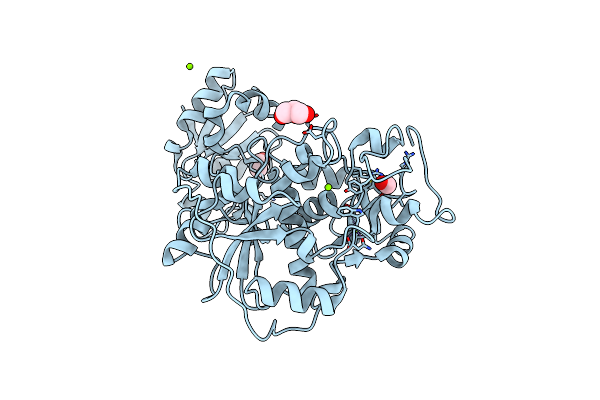 |
Native Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, FMT, EOH, GOL |
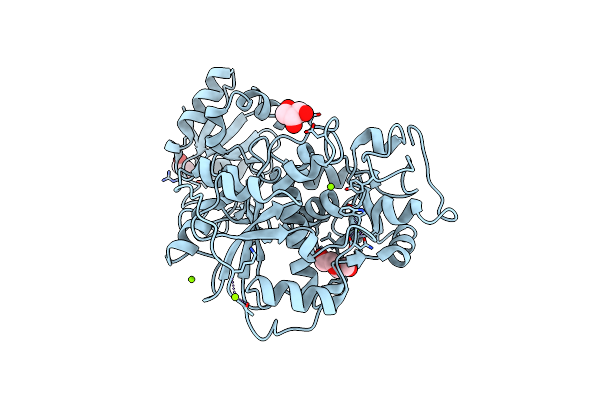 |
Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola In Persulfide Form.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, EDO, PEG, GOL |