Search Count: 4,734
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE |
 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Leca Lectin In Complex With Deuterated Galactose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: RFP |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: MOTOR PROTEIN |
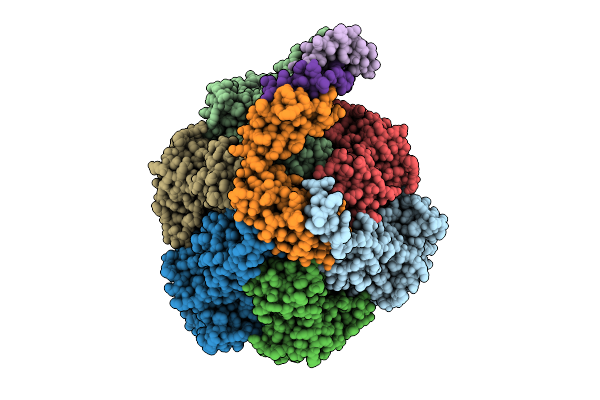 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, PO4 |
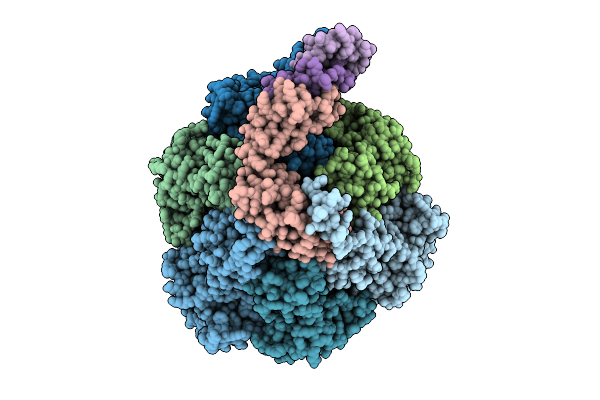 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
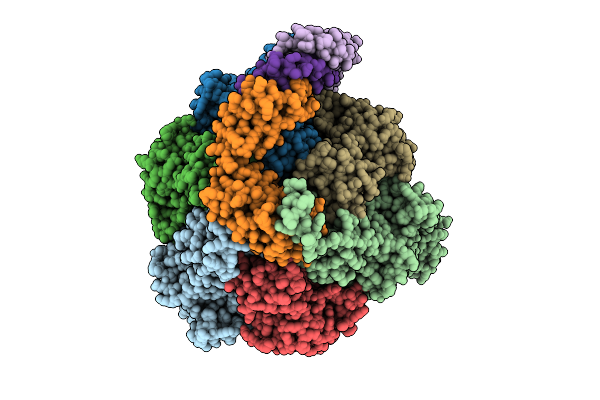 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
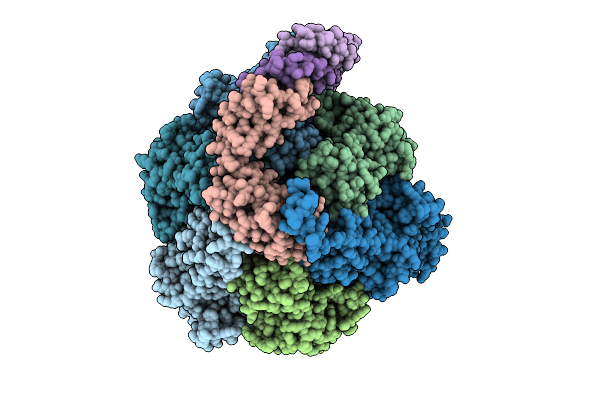 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
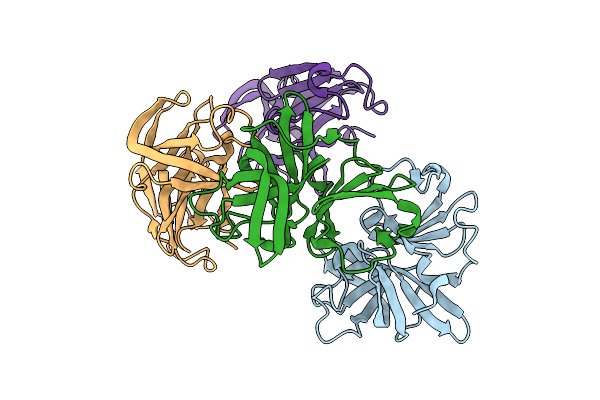 |
Protease From Norovirus Sydney Gii.4 Strain With Crystallization Epitope Mutation H50Y
Organism: Norovirus sydney 2212
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN |
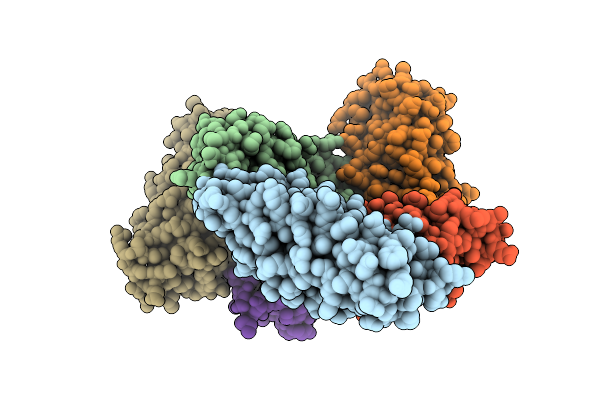 |
Nanoparticle Crystal Structure Of A Thermostabilized Mutant Of An As-Isolated Ftna (Ferritin) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: METAL BINDING PROTEIN |
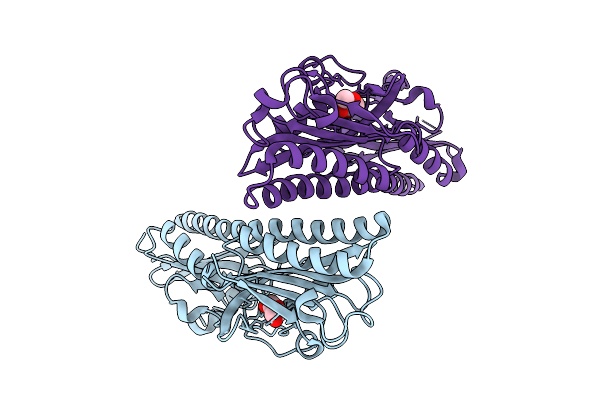 |
Ligand Binding Domain Of Pseudomonas Aeruginosa Pao1 Chemoreceptor Tlpq In Complex With Mhf
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: SIGNALING PROTEIN Ligands: PEG, 4XX |

