Search Count: 64
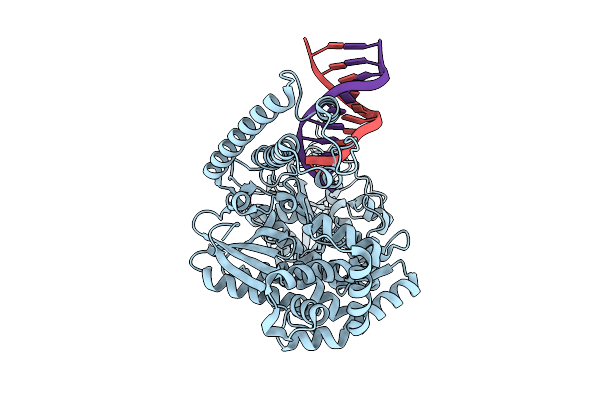 |
Organism: Plasmodium falciparum, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: REPLICATION |
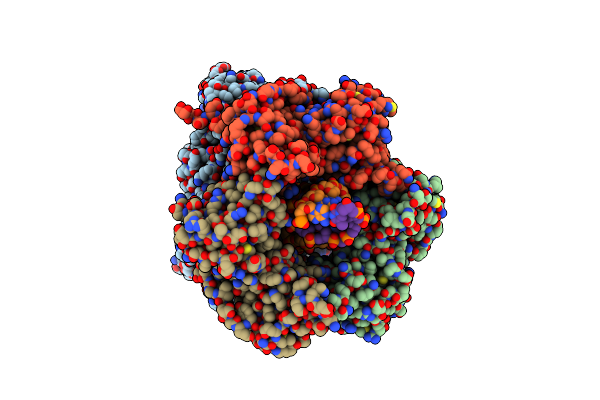 |
Human Polymerase Epsilon Bound To Pcna And Dna In The Nucleotide Exchange State
Organism: Homo sapiens, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: DNA Binding Protein/DNA Ligands: SF4 |
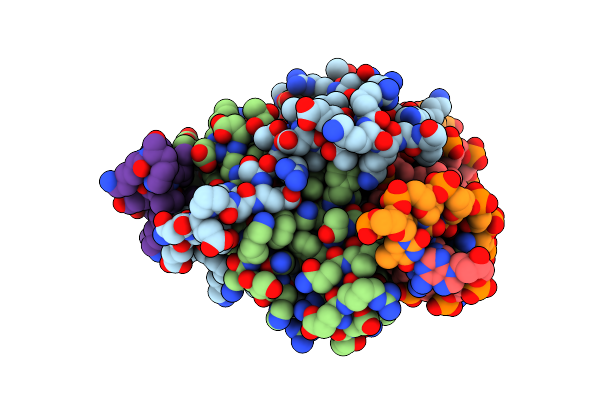 |
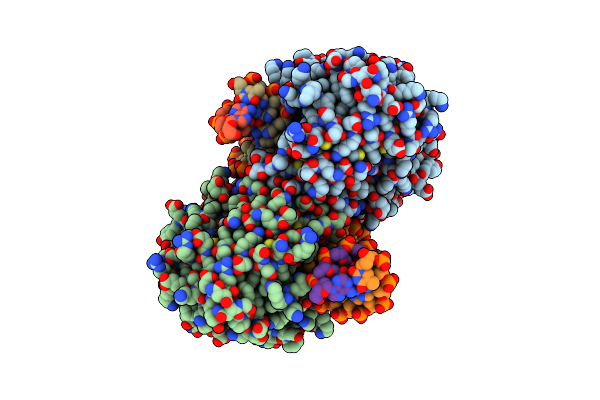
Crystal Structure Of The Mads-Box/Mef2 Domain Of Mef2D Bound To Dsdna And Hdac7 Deacetylase Binding Motif
Organism: Homo sapiens, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: TRANSCRIPTION |
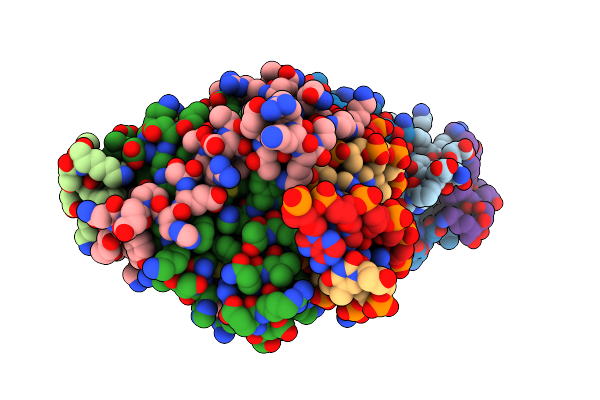 |
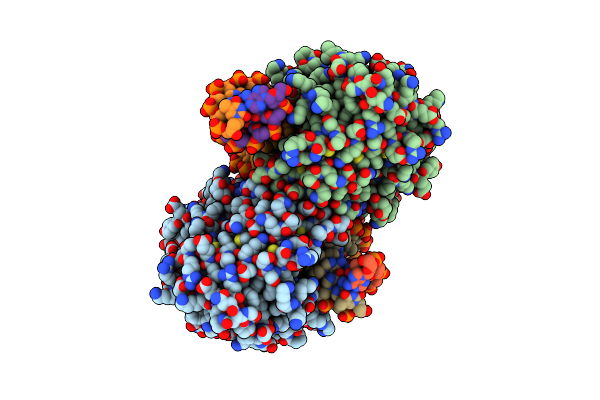
Crystal Structure Of Mads-Box/Mef2D N-Terminal Domain Bound To Dsdna And Hdac9 Deacetylase Binding Motif
Organism: Homo sapiens, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-04-17 Classification: TRANSCRIPTION |
 |
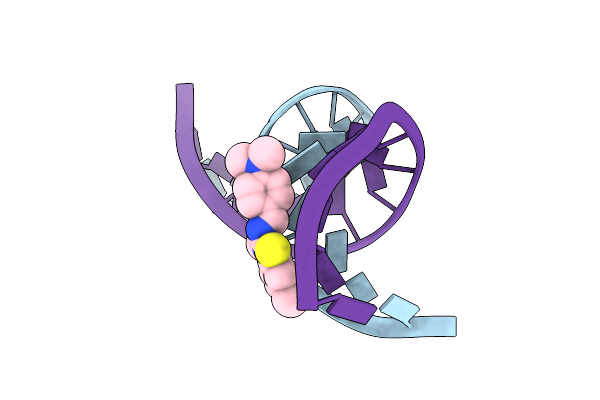
Structure Of Non-Hydrolyzable Atp (Apcpp) Binds To Cyclic Gmp Amp Synthase (Cgas) Through Mn Coordination
Organism: Mus musculus, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-05-03 Classification: Transferase/DNA Ligands: APC, ZN, MN |
 |
Organism: Mus musculus, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-05-03 Classification: Transferase/DNA Ligands: ATP, MG, ZN |
 |
Organism: Arabidopsis thaliana, Dna molecule
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-03-29 Classification: TRANSCRIPTION |
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-01-18 Classification: DNA Ligands: O89 |
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2023-01-18 Classification: DNA |
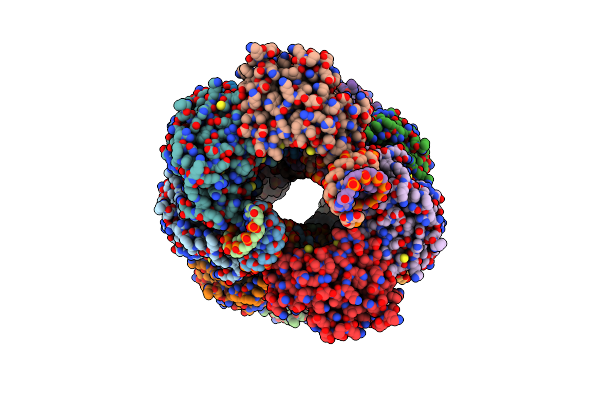 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: HYDROLASE/DNA |
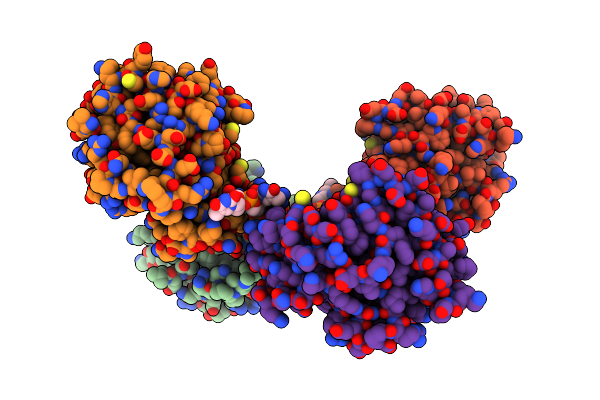 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE/DNA |
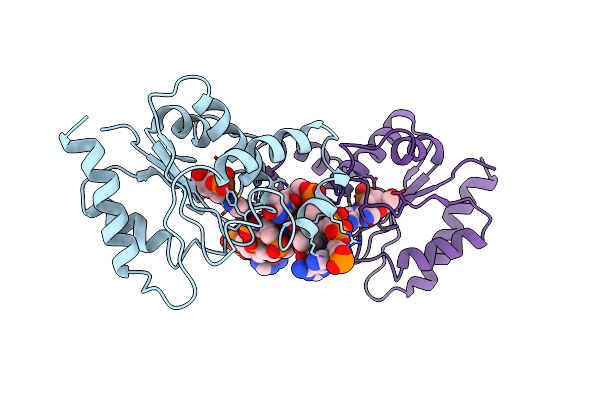 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE/DNA Ligands: ACK |
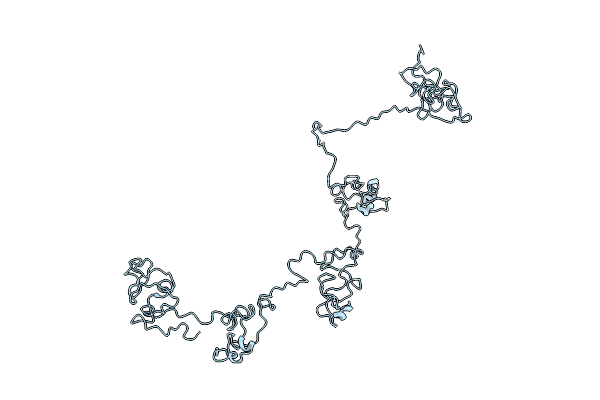 |
Organism: Cylindrotheca fusiformis
Method: SOLUTION NMR Release Date: 2016-12-21 Classification: STRUCTURAL PROTEIN |
 |
Structure Of Nadh-Preferring Ketol-Acid Reductoisomerase From An Uncultured Archean
Organism: Uncultured archaeon gzfos26g2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-04-22 Classification: OXIDOREDUCTASE Ligands: MG, NAI, GOL, HIO |
 |
Structure Of The Pscd4-Domain Of The Cell Wall Protein Pleuralin-1 From The Diatom Cylindrotheca Fusiformis
Organism: Cylindrotheca fusiformis
Method: SOLUTION NMR Release Date: 2015-02-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Silene latifolia subsp. alba
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 1999-10-19 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Organism: Silene latifolia subsp. alba
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-10-15 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
 |
 |

