Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
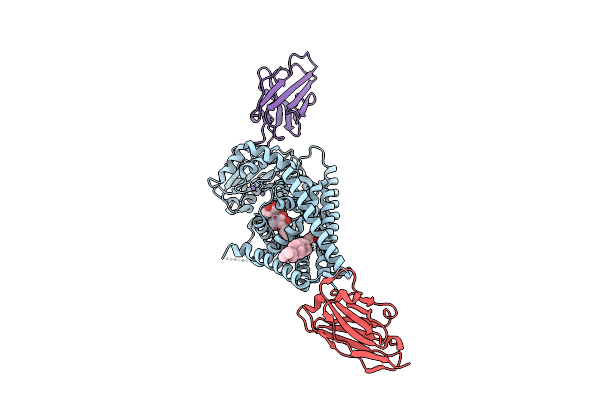 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: Y01, MN, LMT |
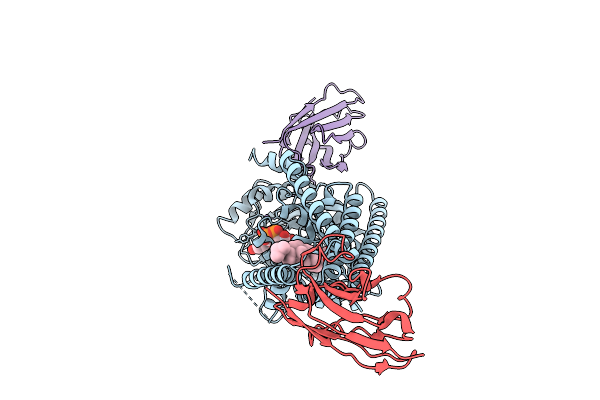 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, MN, Y01 |
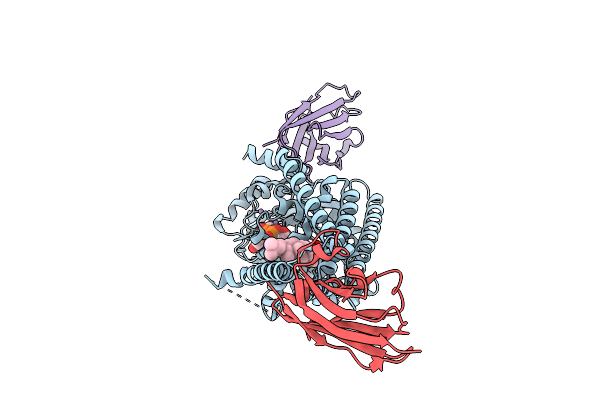 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, Y01, MN |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: Y01, MN |
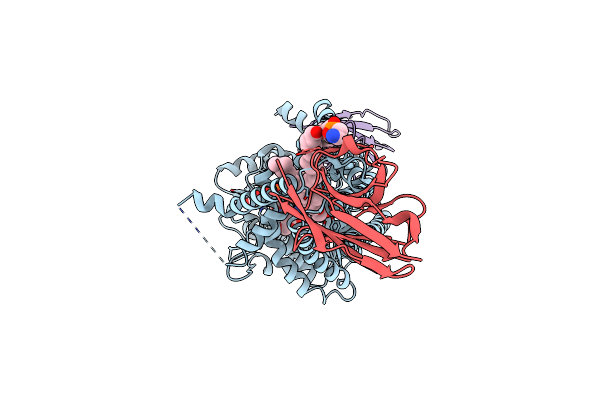 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: 3PE, NAG, UGA, Y01, MN |
 |
Chlorella Virus Hyaluronan Synthase Bound To Glca Extended Glcnac Primer And Udp
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: UDP, Y01, 3PE, MN |
 |
High-Resolution Crystal Structure Of The Mu8.1 Conotoxin From Conus Mucronatus
Organism: Conus mucronatus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-08-23 Classification: TOXIN Ligands: MES, P4G, BCT, SO4, ZN, NA |
 |
Organism: Conus mucronatus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-11-16 Classification: TOXIN Ligands: EPE |
 |
Organism: Conus mucronatus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-11-02 Classification: TOXIN Ligands: IOD, CD |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: Y01, MN, 3PE |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: 3PE, Y01, UDP, MN |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: UD1, 3PE, Y01, MN |
 |
Chlorella Virus Hyaluronan Synthase In The Glcnac-Primed Channel-Closed State
Organism: Lama glama, Paramecium bursaria chlorella virus cz-2
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Chlorella Virus Hyaluronan Synthase In The Glcnac-Primed, Channel-Open State
Organism: Lama glama, Paramecium bursaria chlorella virus cz-2
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01, 3PE |
 |
Crystal Structure Of The Gracilariopsis Lemaneiformis Alpha-1,4- Glucan Lyase Covalent Intermediate Complex With 5-Fluoro-Idosyl- Fluoride
Organism: Gracilariopsis lemaneiformis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-03-27 Classification: LYASE Ligands: B9D, GOL, 5DI |
 |
Crystal Structure Of The Gracilariopsis Lemaneiformis Alpha-1,4- Glucan Lyase Covalent Intermediate Complex With 5-Fluoro-Glucosyl- Fluoride
Organism: Gracilariopsis lemaneiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-03-27 Classification: LYASE Ligands: 5GF, GOL, AFR |
 |
Crystal Structure Of The Gracilariopsis Lemaneiformis Alpha-1,4- Glucan Lyase
Organism: Gracilariopsis lemaneiformis
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2011-01-19 Classification: LYASE Ligands: GOL, ACT, CL |
 |
Crystal Structure Of The Gracilariopsis Lemaneiformis Alpha-1,4- Glucan Lyase With Acarbose
Organism: Gracilariopsis lemaneiformis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-01-19 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of The Gracilariopsis Lemaneiformis Alpha- 1,4-Glucan Lyase With Deoxynojirimycin
Organism: Gracilariopsis lemaneiformis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-01-19 Classification: LYASE Ligands: NOJ, GOL, CL |
 |