Search Count: 611
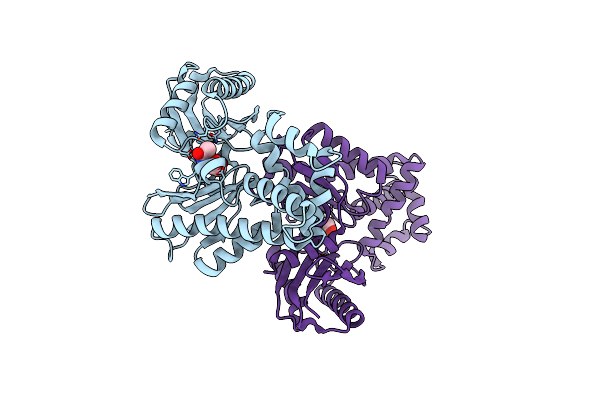 |
The Crystal Structures Of Murk In Complex With N-Acetylglucosamine From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: NAG |
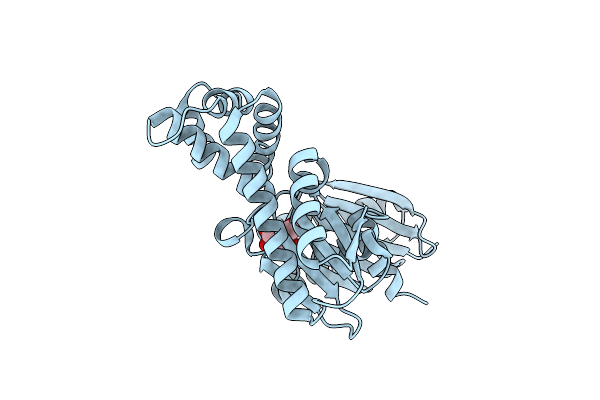 |
The Crystal Structures Of Murk In Complex With Glucose From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: BGC |
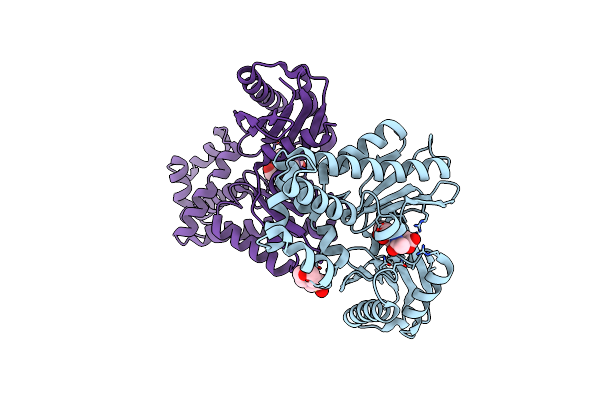 |
The Crystal Structures Of Murk In Complex With N-Acetylmuramic Acid (Murnac) From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: AMU, MES |
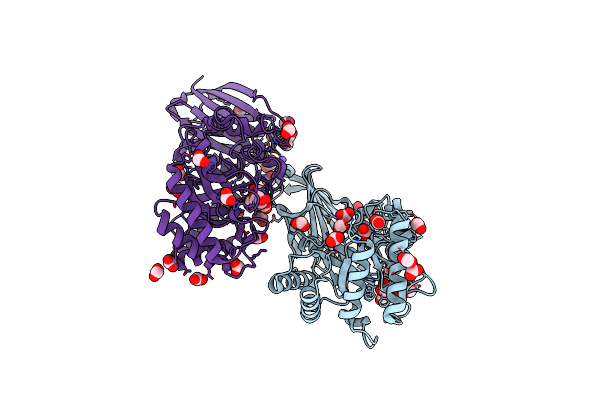 |
A X-Ray Crystallographic Structure Model Of A Glycoside Hydrolase (Gh) Family 39 (Gh39)-Like Enzyme Encoded Within A Xylan Utilization Locus Of The Psol1 Megaplasmid Of Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-11-13 Classification: HYDROLASE Ligands: GOL, FMT, PGE, BCN, NA |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: EDO, CL |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: HYDROLASE |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of A Thioredoxin From Clostridium Acetobutylicum At 1.75 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.3 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, PEG, ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.9 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, PEG, ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.64 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PEG |
 |
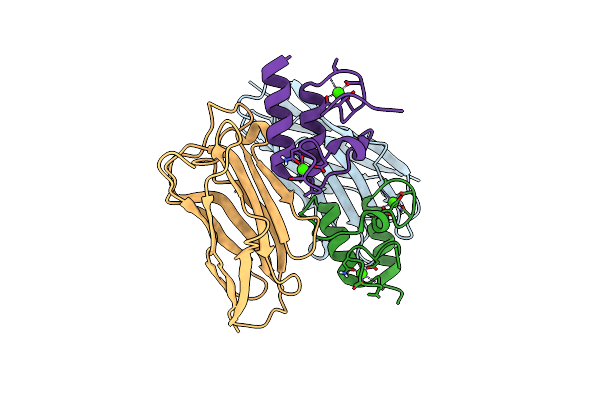
Crystal Structure Of The Complex Of A Ferredoxin-Flavin Thioredoxin Reductase And A Thioredoxin From Clostridium Acetobutylicum At 2.9 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, EDO |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coenzymea Dehydrogenase From Clostridium Acetobutylicum Complexed With Nad+
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase From Clostridium Acetobutylicum, Apo Form
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE |
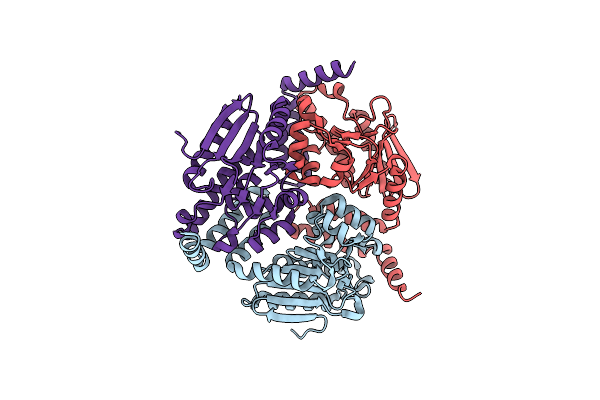 |
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-02-14 Classification: LYASE |
 |
X-Ray Crystallographic Protein Structure Of The Glycoside Hydrolase Family 30 Subfamily 8 Xylanase, Xyn30A, From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-08-10 Classification: HYDROLASE |

