Search Count: 5,003
 |
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: 1AX |
 |
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: CELL ADHESION |
 |
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: CELL ADHESION Ligands: IOD |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 353K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: PO4, ADE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 323K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: PEG, PO4, MTA |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 293K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: PEG, PO4, MTA |
 |
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL TRANSPORT Ligands: PGE, PEG, CL |
 |
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL TRANSPORT Ligands: FCE, FE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 333K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSFERASE Ligands: PO4, MTA |
 |
Crystal Structure Of Peni Beta-Lactamase From Burkholderia Pseudomallei Complex With Taniborbactam
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: KJK, GOL |
 |
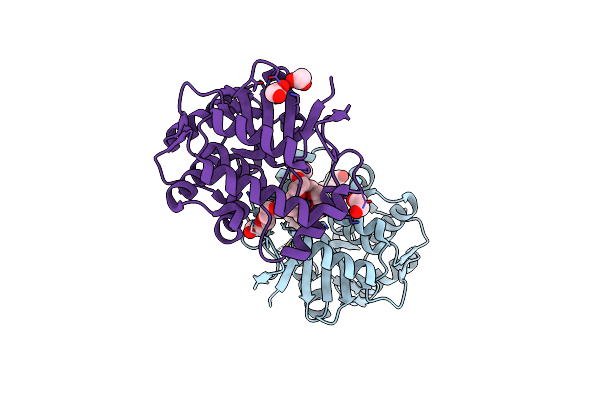
Crystal Structure Of Bf3526 Peptidase From Bacteroides Fragilis In Complex With A Peptide
Organism: Bacteroides fragilis nctc 9343, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: K, ZN, EDO, POL, PEG, BU1, HEZ |
 |
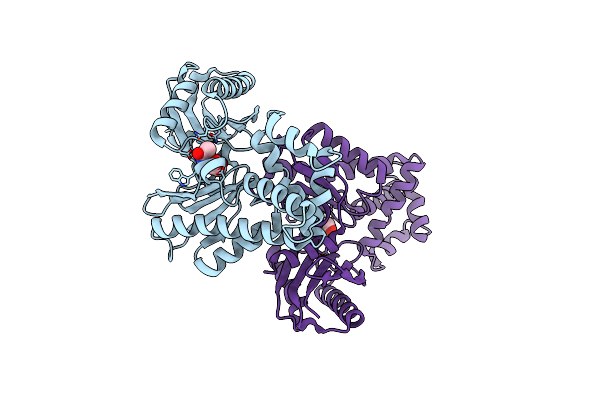
Crystal Structure Of The Surface Protein (Cd630_07380) From Clostridium Difficile Strain 630
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
 |
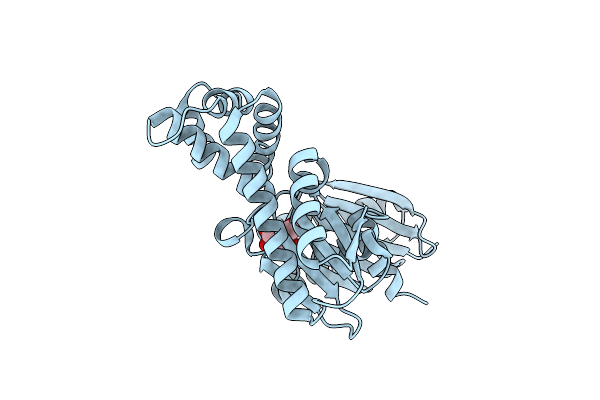
The Crystal Structures Of Murk In Complex With N-Acetylglucosamine From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: NAG |
 |
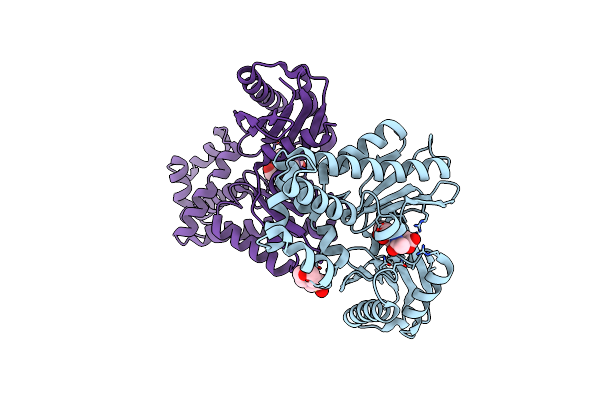
The Crystal Structures Of Murk In Complex With Glucose From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: BGC |
 |
The Crystal Structures Of Murk In Complex With N-Acetylmuramic Acid (Murnac) From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: AMU, MES |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168, Geobacillus thermodenitrificans ng80-2, Lama glama, Haloarcula marismortui atcc 43049
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: PROTEIN TRANSPORT Ligands: MG, BEF, ADP |
 |
Organism: Escherichia coli k-12, Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-02-12 Classification: ANTITOXIN Ligands: GOL |
 |
Crystal Structure Of The Spore Gernation Lytic Transglycosylase Slec From Clostridioides Difficile In Its Zymogenic Form (Prepro-Slec)
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
Crystal Structure Of Hemolysin Co-Regulated Protein 1 (Hcp1) Variantb From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-12-25 Classification: PROTEIN TRANSPORT Ligands: SO4 |

