Search Count: 3,787
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
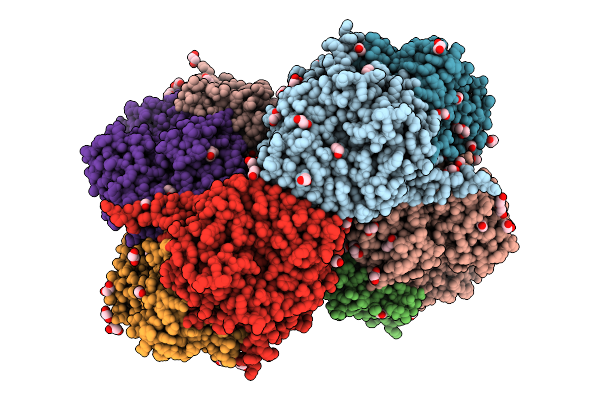
Crystal Structure Of Aetf-L183F/V220I/S523A In Complex With Fad And L-Tryptophan
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: TRP, FAD |
 |
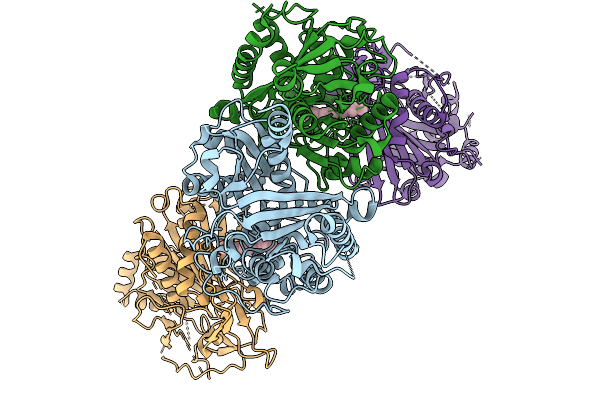
Organism: Azotobacter vinelandii dj
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q, PEG, EDO, 1PE, PGE, MG, PG4 |
 |
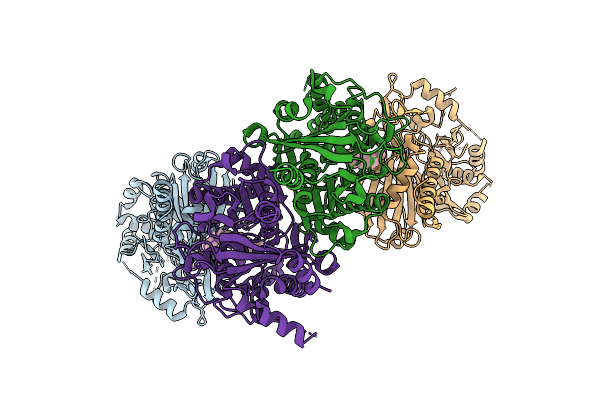
Organism: Citrus sinensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PLANT PROTEIN Ligands: D12 |
 |
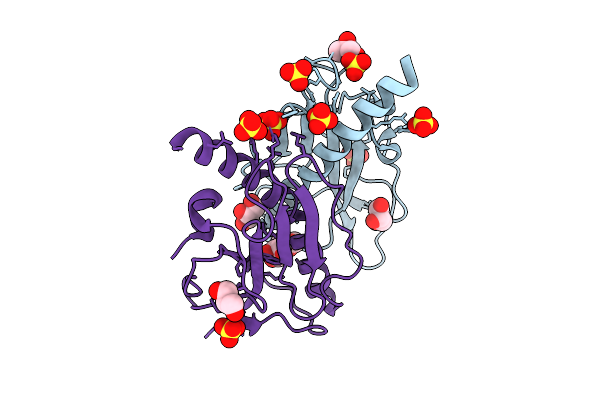
Organism: Citrus sinensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PLANT PROTEIN Ligands: D12, R16 |
 |
Organism: Dissulfurispira thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Citrus sinensis, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: PLANT PROTEIN Ligands: A1IRJ, MN, CL |
 |
Organism: Citrus sinensis, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: PLANT PROTEIN Ligands: A1IRN, MN, CL |
 |
X-Ray Crystal Structure Of The Cspyl1 5M (V112L, T135L, F137I, T153I, V168A)-Icb-Hab1 Ternary Complex
Organism: Citrus sinensis, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: PLANT PROTEIN Ligands: A1IRN, MN, CL |
 |
Crystal Structure Of Uracil-Dna Glycosylase From Burkholderia Thailandensis
Organism: Burkholderia thailandensis e264
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: CL, SO4 |
 |
Organism: Candidatus odinarchaeota
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: CELL CYCLE Ligands: G2P |
 |
Organism: Burkholderia thailandensis e264
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: CELL ADHESION Ligands: GOL, SO4, BR |
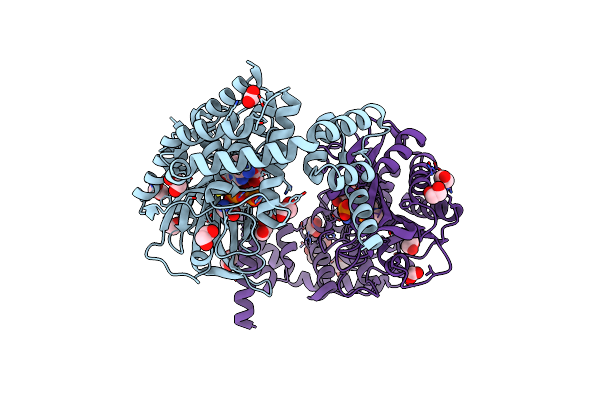 |
Crystal Structure Of Alanyl-Trna Synthetase In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, EDO, FMT, MLT, CIT |
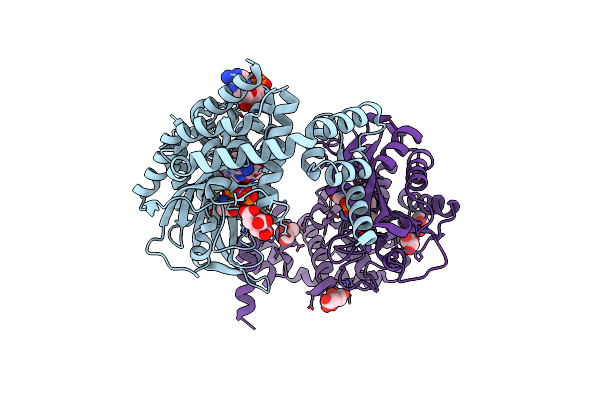 |
Crystal Structure Of Alanyl-Trna Synthetase L219M Mutant In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, CIT, TRS, GOL |
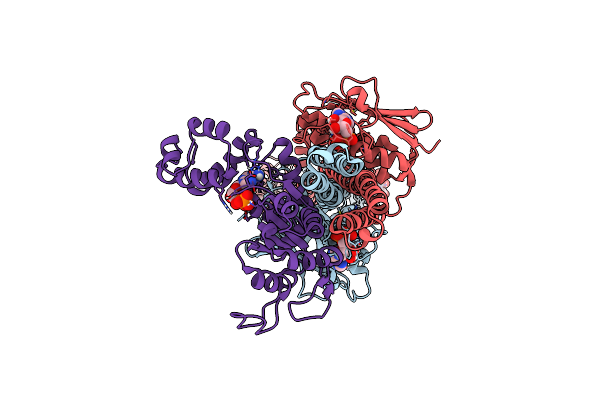 |
Organism: Azotobacter vinelandii dj
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: GENE REGULATION Ligands: FAD, ADP |
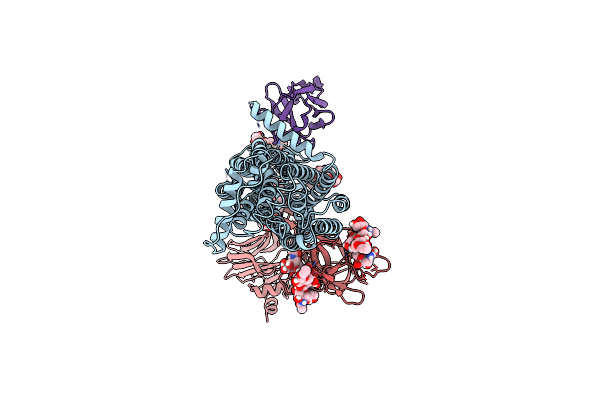 |
Cryo-Em Structure Of Native Sv2A In Complex With Tent-Hc, Gangliosides And Pro-Macrobody 5
Organism: Clostridium tetani e88, Lama glama, Escherichia coli (strain k12), Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: NAG, 6UZ, A1IHM, A1IHN |
 |
Cryo-Em Structure Of Native Sv2A In Complex With Tent-Hc, Pro-Macrobody 5 And Levetiracetam
Organism: Clostridium tetani e88, Lama glama, Escherichia coli k-12, Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: NAG, 6UZ, UKX |
 |
Organism: Spiromastix sp. scsio f190
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-02-12 Classification: BIOSYNTHETIC PROTEIN |

