Search Count: 5,415
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
 |
Crystal Structure Of Sars-Cov-2 Papain-Like Protease (Cys111Ser) In Complex With Yl1004
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EOE, ZN, CL, PEG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Crystal Structure Of Sars-Cov-2 3Cl Protease (3Clpro) In Complex With Compound 8
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-050-22 At 1.16 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1C3L |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Inhibitor Grl-062-22 At 1.65 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1C3M, NA |
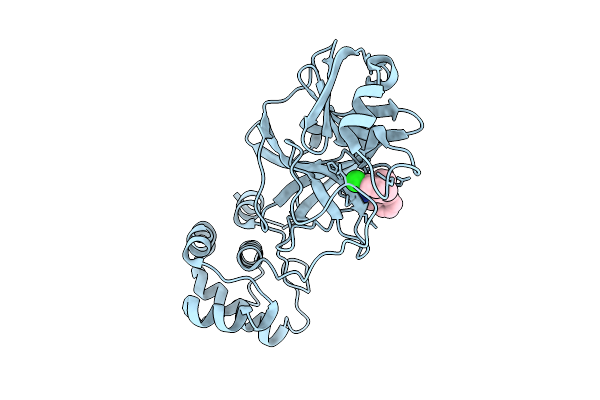 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Iso-Quinoline-Derived Inhibitor Fd6-31
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1EH0 |
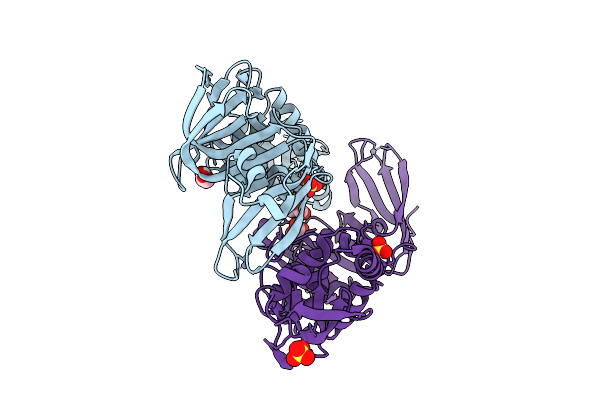 |
A Rare Open Conformation For Ubl2 Domain Of Papain-Like Protease C111S Of Sars-Cov2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: GOL, ZN, SO4 |
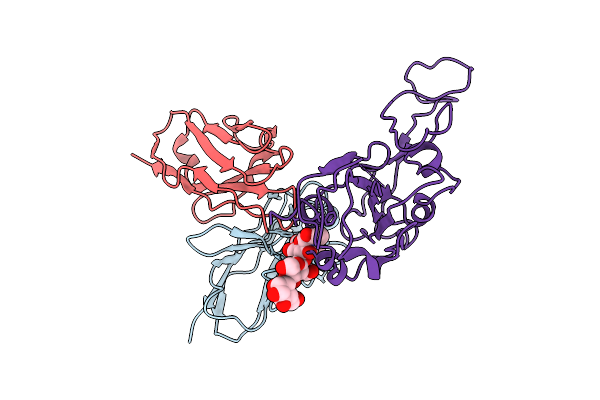 |
Cryo-Em Structure Of Sars-Cov-2 Ba.5 Spike Protein In Complex With Nab 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
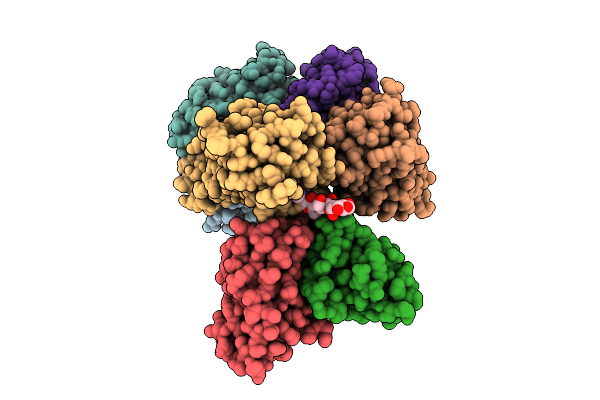 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Three-Nab 8H12, 3E2 And 1C4
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.1 Spike Protein In Complex With Three-Nab 8H12, 3E2 And 1C4
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.2 Spike Protein In Complex With Triple-Nab 8H12, 3E2 And 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

