Search Count: 31
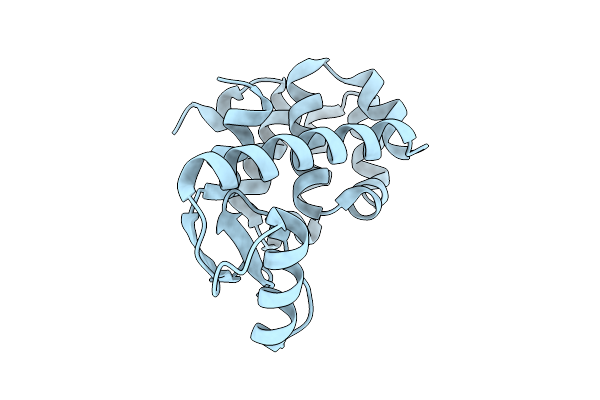 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A (Apo State)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (1 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (10 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND, NA |
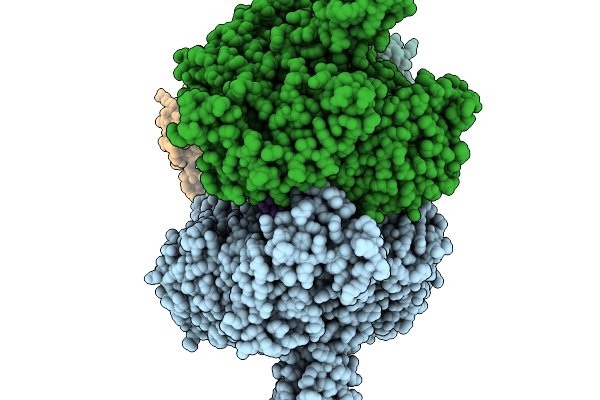 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 1
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: replication/DNA |
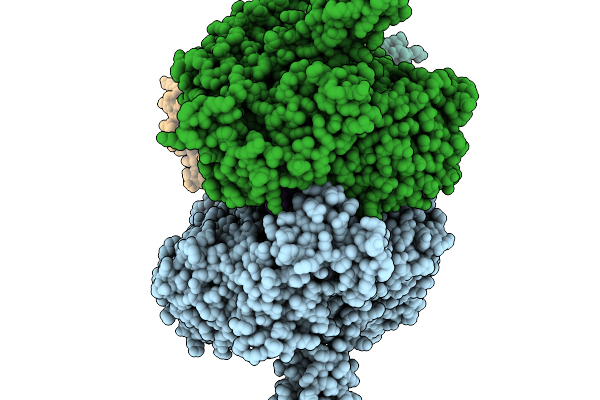 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 2
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: replication/DNA |
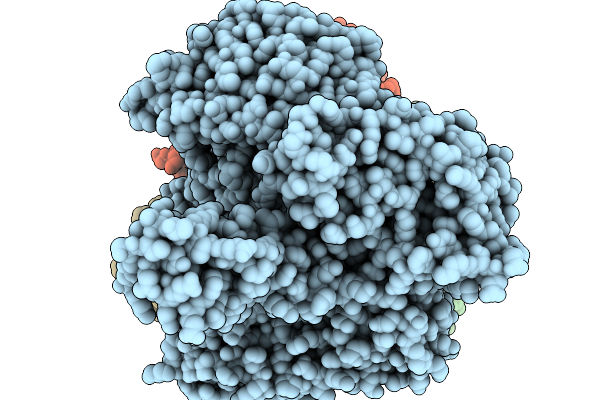 |
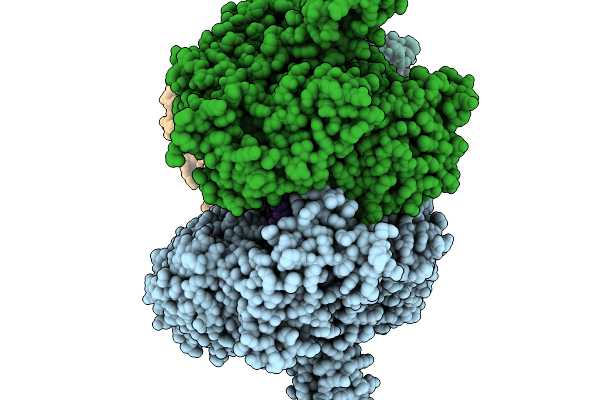
T4 Bacteriophage Replicative Holoenzyme Containing Triple Mutations D75R, Q430E, And K432E In The Exonuclease-Deficient Polymerase
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: Transferase/DNA |
 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 3
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: Replication/DNA Ligands: CA, D3T |
 |
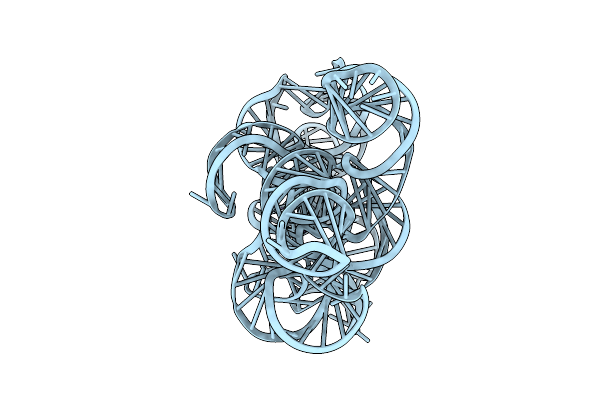
Cryo-Em Structure Of Linear Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
 |
Cryo-Em Structure Of Circular Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
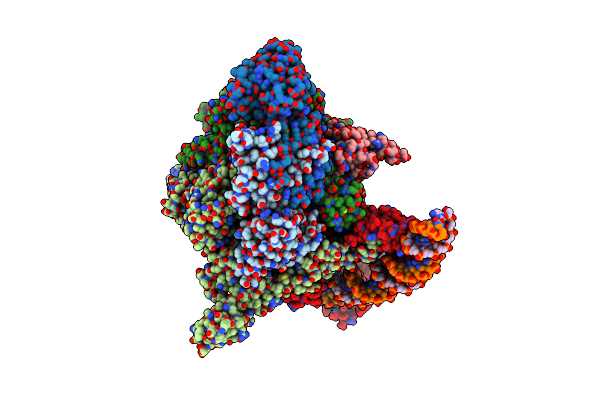 |
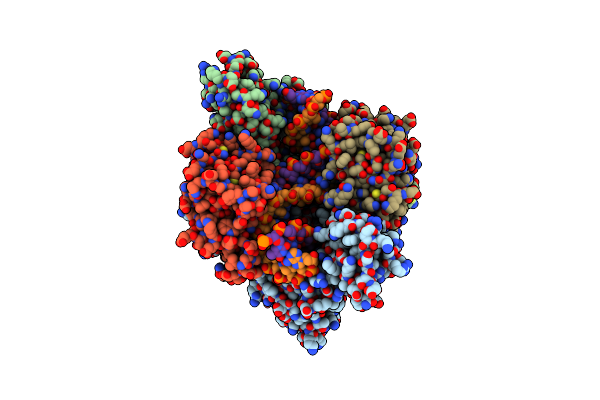
Organism: Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: TRANSCRIPTION/DNA/RNA |
 |
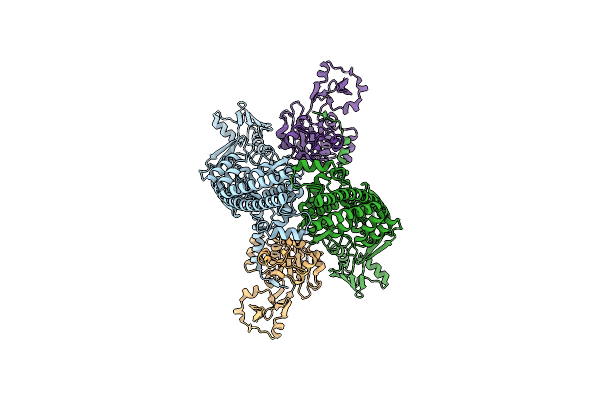
Organism: Escherichia phage t4, Salmonella phage chi, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE Ligands: MG |
 |
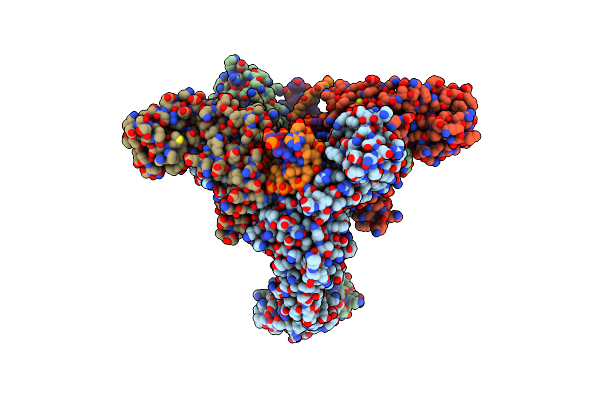
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE |
 |
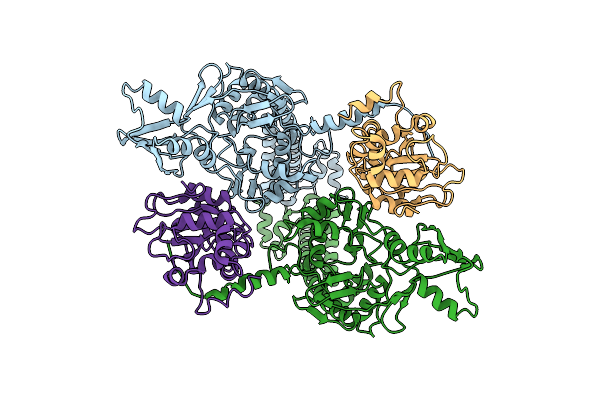
Organism: Escherichia phage t4, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE Ligands: MG |
 |
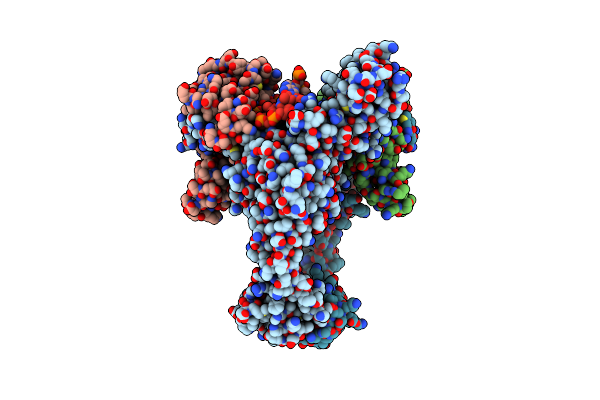
Organism: Escherichia phage t4, Enterobacteria phage t6
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE |
 |
Structure Of Phage T6 Topoisomerase Ii Central Domain Bound With Dna And M-Amsa
Organism: Escherichia phage t4, Enterobacteria phage t6, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE Ligands: MG, ASW |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE |
 |
Organism: Escherichia phage t4, Enterobacteria phage t6
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE Ligands: ANP |
 |
Organism: Escherichia phage t4, Enterobacteria phage t6, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: ISOMERASE Ligands: ANP |
 |
Organism: Escherichia coli (strain k12), Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA |
 |
Organism: Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA |

