Search Count: 1,004
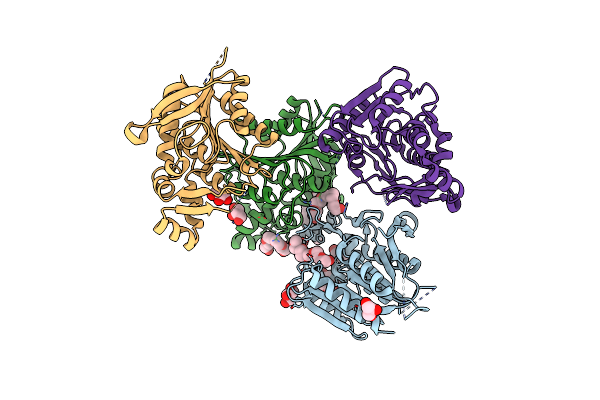 |
Crystal Structure Of Sm0281, An Alpha/Beta Hydrolase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: GOL, PE3, CL |
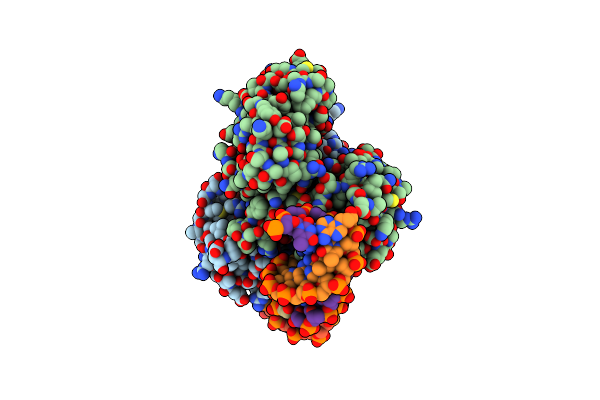 |
Organism: Sinorhizobium meliloti 1021, Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: CMP, MG |
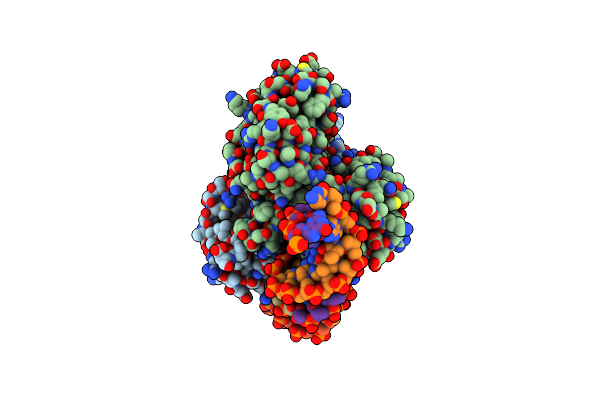 |
Organism: Sinorhizobium meliloti 1021, Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: PCG |
 |
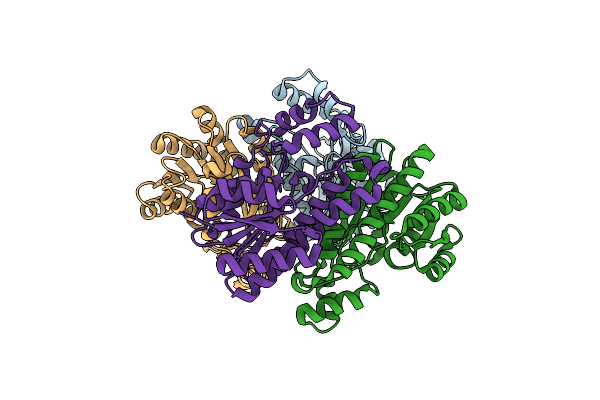
Nmr Structure Of The Zeta-Subunit Of The F1F0-Atpase From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti 1021
Method: SOLUTION NMR Release Date: 2021-10-13 Classification: PROTEIN BINDING |
 |
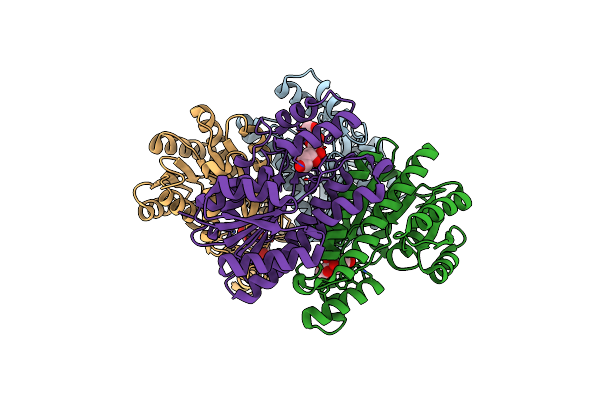
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-06-24 Classification: OXIDOREDUCTASE |
 |
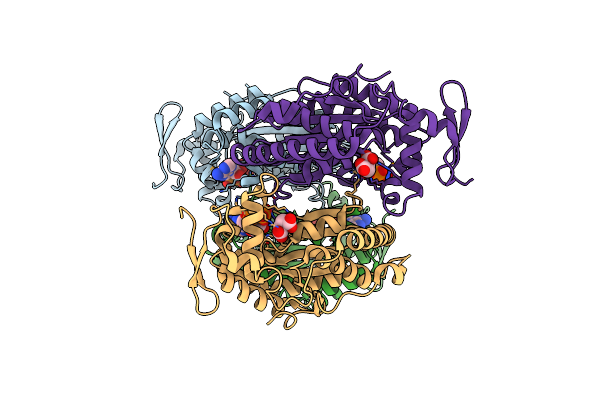
Structure Of Sorbitol Dehydrogenase From Sinorhizobium Meliloti 1021 Bound To Sorbitol
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-24 Classification: OXIDOREDUCTASE Ligands: SOR |
 |
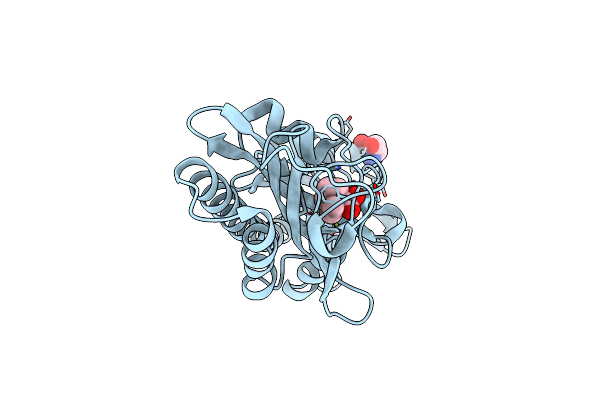
Crystal Structure Of Polyphosphate Kinase 2 Class I (Smc02148) In Complex With Adp
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-07-10 Classification: TRANSFERASE Ligands: ADP, MLT, AMP |
 |
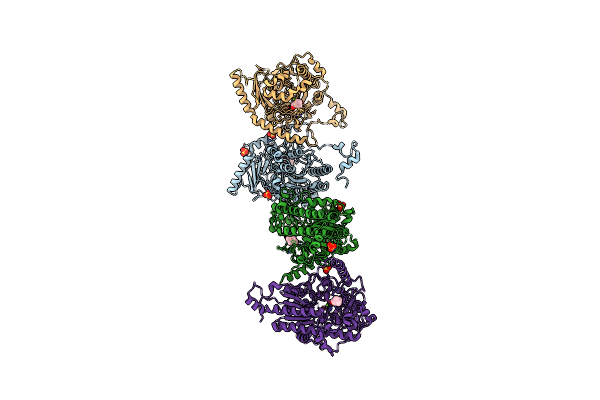
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-17 Classification: MEMBRANE PROTEIN Ligands: TRS, PBE |
 |
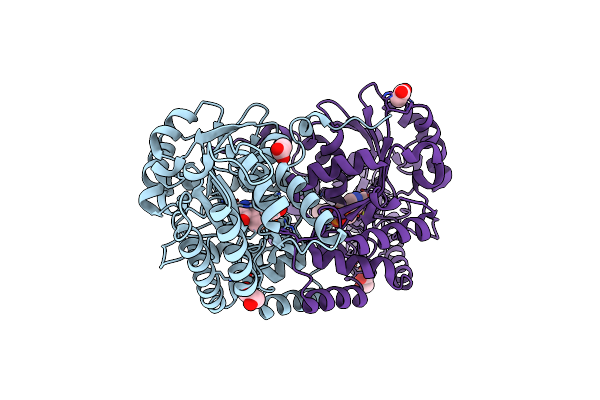
Choline Sulfatase From Ensifer (Sinorhizobium) Meliloti Cocrystalized With Choline
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: CHT, MG, SO4 |
 |
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PLP, ACT, GOL |
 |
Crystal Structure Of The Prephenate Aminotransferase From Rhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PLP |
 |
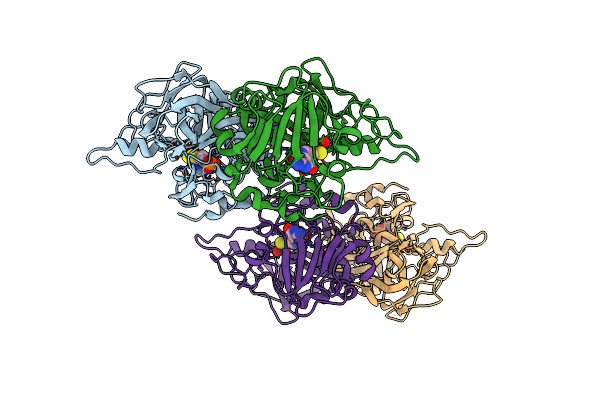
Organism: sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-02-28 Classification: HYDROLASE Ligands: CA |
 |
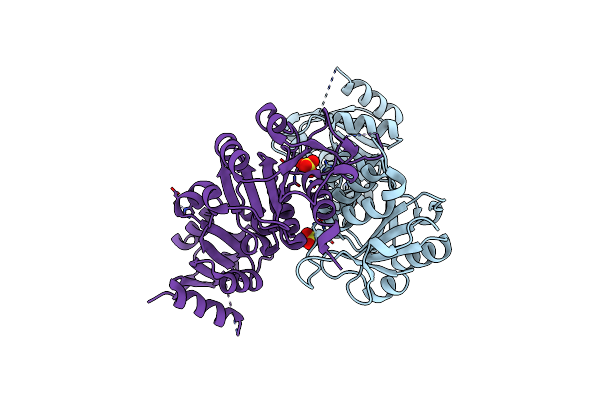
Crystal Structure Of The Sulfite Dehydrogenase, Sort R78Q Mutant From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: MSS |
 |
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2017-08-02 Classification: RNA BINDING PROTEIN Ligands: CO, SO4 |
 |
Crystal Structure Of Adomet Bound Rrna Methyltransferase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-08-02 Classification: TRANSFERASE Ligands: CO, SO4, SAM |
 |
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-05-31 Classification: TRANSCRIPTION |
 |
Crystal Structure Of The Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-05-24 Classification: OXIDOREDUCTASE Ligands: MSS, GOL |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And Malonate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, MLA, NA |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Citrate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: OXL, NDP, GOL, CL, NA |

