Search Count: 5,562
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Atp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ATP, PEG, MG, PGE, GOL, ADP, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Adp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ADP, PEG, MG, PGE, GOL, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Amp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: AMP, PGE, MG, PEG, PG4, GOL |
 |
Crystal Structure Of Ornithine Carbamoyltransferase From Burkholderia Xenovorans
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: CL |
 |
Crystal Structure Of Ornithine Carbamoyltransferase From Burkholderia Xenovorans In Complex With Phosphono Carbamate
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: CP, PO4, CL |
 |
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: GAL, PG4, GOL, PO4, CA, SO4 |
 |
Crystal Structure Of A Galactose Oxidase From Pseudoarthrobacter Siccitolerans
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: GOL, PG4, CA, CL, BR |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Complex Of Fapn With Fcov-23 Rbd
Organism: Felis catus, Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Fcov-23 S Short
Organism: Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN Ligands: NAG, PAM |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Fcov-23 S Do In Proximal Conformation (Local Refinement)
Organism: Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Fcov-23 S Long With Do In Swung-Out Conformation
Organism: Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN Ligands: NAG, PAM |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Fcov-23 S Long Domain 0 In Swung-Out Conformation (Local Refinement)
Organism: Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
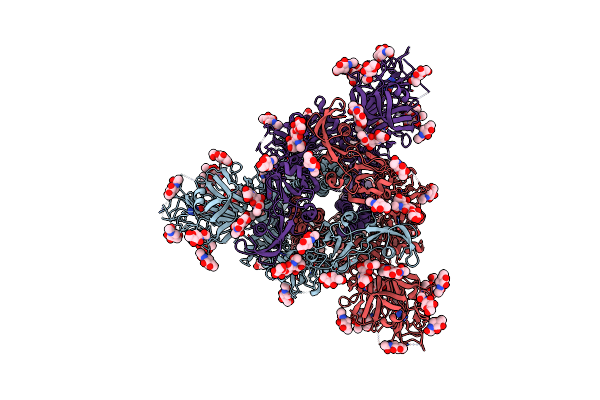
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Fcov-23 S Long With Do In Mixed Conformations (Global Refinement).
Organism: Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN Ligands: NAG, PAM |
 |
Cryo-Em Structure Of Cn-Hedgehogcov (Hku31/Erinaceus Amurensis/China/2014) S-Trimer In A Locked-2 Conformation
Organism: Erinaceus hedgehog coronavirus hku31, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: FOL, NAG |
 |
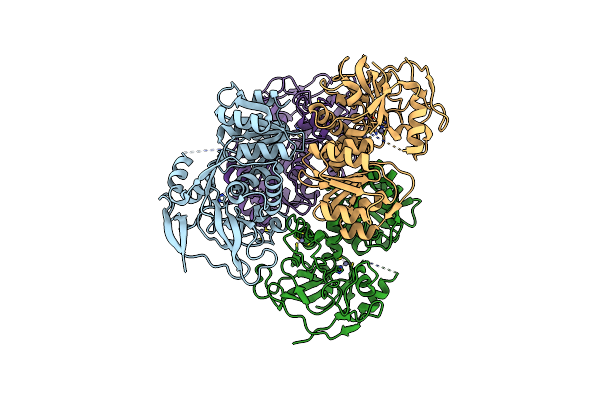
Cryo-Em Structure Of Cn-Hedgehogcov (Hku31/Erinaceus Amurensis/China/2014) S-Trimer In A Locked-1 Conformation
Organism: Erinaceus hedgehog coronavirus hku31, Tequatrovirus t4
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: NAG, FOL, EIC |
 |
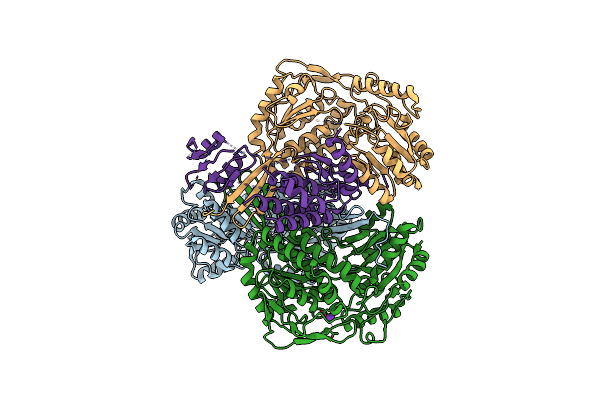
Organism: Paracoccus denitrificans
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
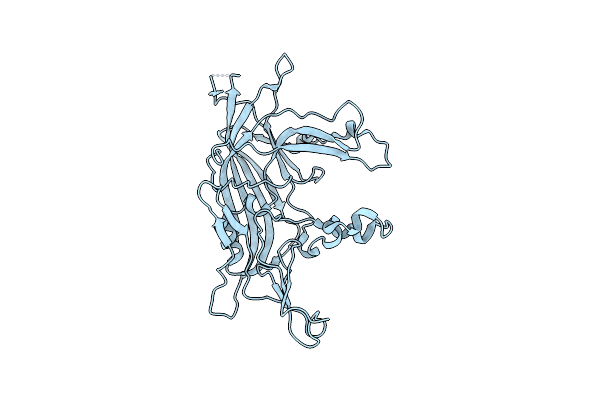
Organism: Paracoccus denitrificans
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: K |
 |
Capsid Structure Of The Syngnathus Scovelli Chapparvovirus Virus-Like Particle
Organism: Syngnathus scovelli chapparvovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRUS LIKE PARTICLE |
 |
Crystal Structure Of Sm0281, An Alpha/Beta Hydrolase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: GOL, PE3, CL |
 |
Cryo-Em Structure Of Uropathogenic Escherichia Coli Cysk:Cdia:Trna Complex A
Organism: Escherichia coli, Escherichia coli 536
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: TOXIN Ligands: MG |

