Search Count: 498
 |
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: EDO, ZN |
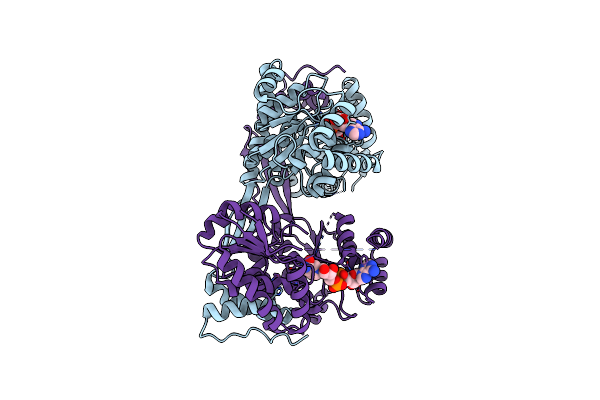 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI |
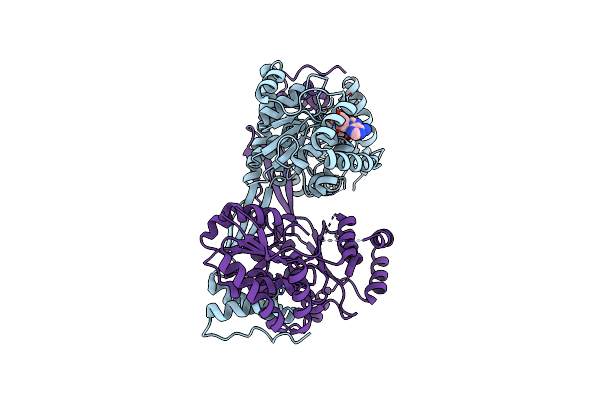 |
Crystal Structure Of Hpsn D352A Mutant From Cupriavidus Pinatubonensis In Complex With Nad+
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
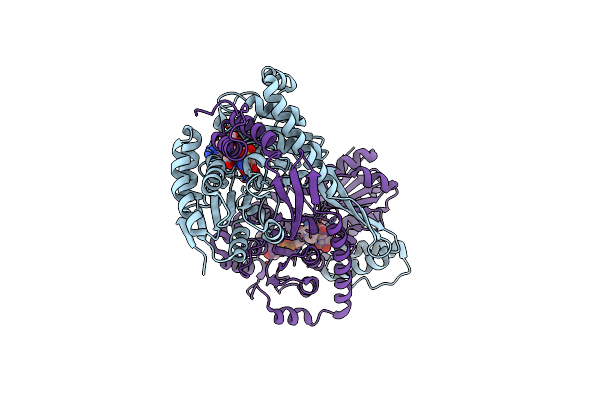 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product Analogue (L-Cysteate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, OCS |
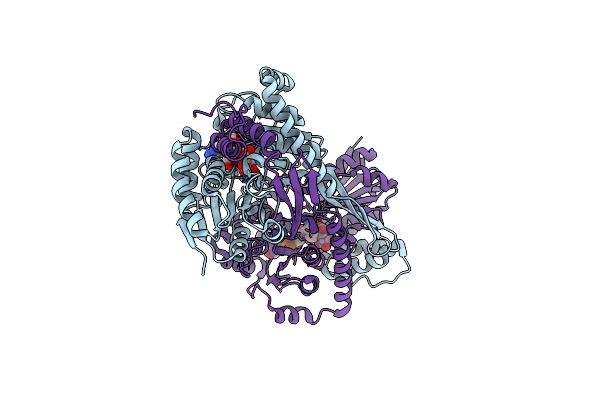 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product (R-Sulfolactate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, 3SL |
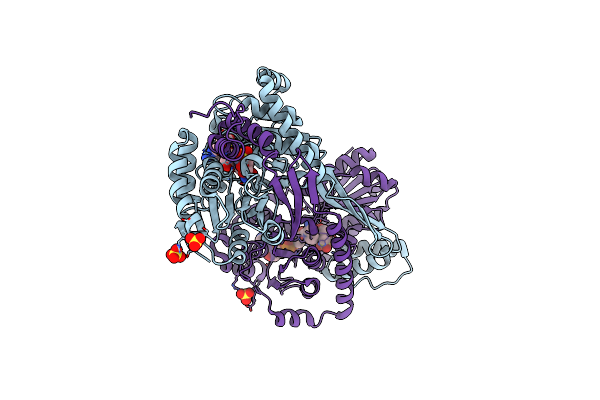 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn H319A Variant From Cupriavidus Pinatubonensis In Complex With Substrate (R-Dhps) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, A1AZH, SO4 |
 |
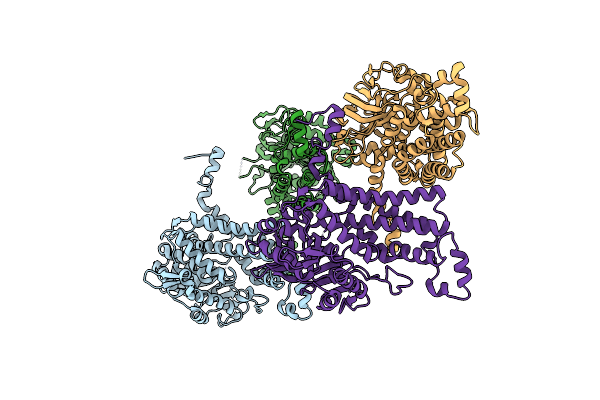
Crystal Structure Of A Putative Cysteine Dioxygnase From Ralstonia Eutropha: An Alternative Modeling Of 2Gm6 From Jcsg Target 361076
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FE, OXY, SO4, EDO |
 |
Organism: Cupriavidus necator jmp134
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-09-19 Classification: OXIDOREDUCTASE |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, GOL, SO4, NAD |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, ISP, SO4, NAD, FU2 |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, FU2, SO4, NAD |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, FU2, SO4, NAD |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-04-25 Classification: OXIDOREDUCTASE Ligands: ZN, P6G |
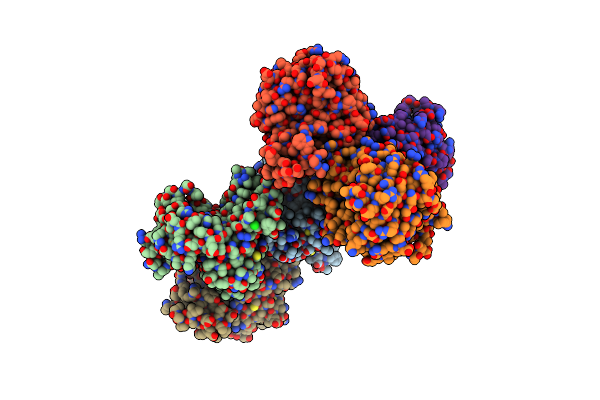 |
Crystal Structure Of A Putative Lyase (Reut_B4148) From Ralstonia Eutropha Jmp134 At 2.44 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2011-03-02 Classification: LYASE Ligands: CL |
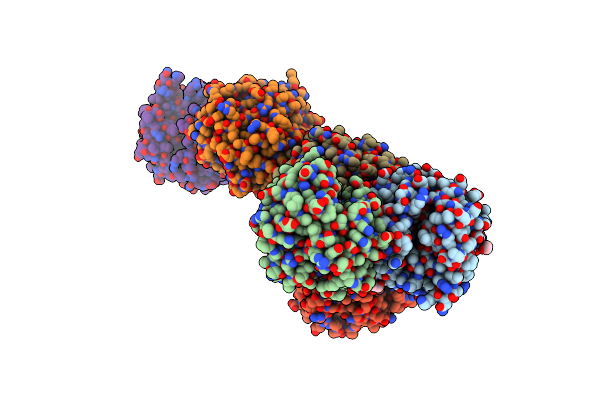 |
Crystal Structure Of A Pfam Duf849 Domain Containing Protein (Reut_A1631) From Ralstonia Eutropha Jmp134 At 1.90 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-08-25 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN, EDO, ACT |
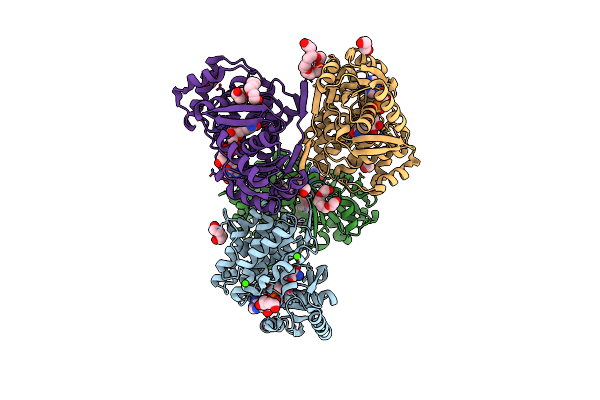 |
Crystal Structure Of Putative Alcohol Dehedrogenase (Yp_298327.1) From Ralstonia Eutropha Jmp134 At 2.10 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-10-06 Classification: OXIDOREDUCTASE Ligands: NAD, CA, CL, PEG, PG4, P6G, GOL, PGE |
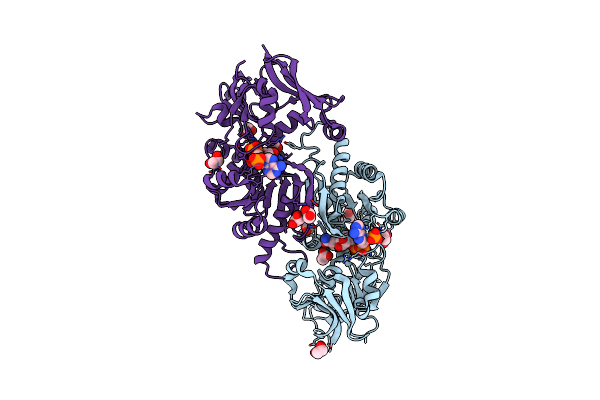 |
Crystal Structure Of Putative Nadph:Quinone Oxidoreductase (Yp_296108.1) From Ralstonia Eutropha Jmp134 At 1.70 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-09-15 Classification: OXIDOREDUCTASE Ligands: NDP, EDO, ACT |
 |
Crystal Structure Of Putative Glyoxalase Superfamily Protein (Yp_299723.1) From Ralstonia Eutropha Jmp134 At 1.90 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-09-08 Classification: LYASE Ligands: P6G, PGE |
 |
Crystal Structure Of Dna Binding Protein (Yp_298823.1) From Ralstonia Eutropha Jmp134 At 1.92 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2009-08-11 Classification: TRANSCRIPTION REGULATOR Ligands: CL, GOL |
 |
Crystal Structure Of Pane/Apba Family Ketopantoate Reductase (Yp_299159.1) From Ralstonia Eutropha Jmp134 At 2.15 A Resolution
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-06-30 Classification: OXIDOREDUCTASE Ligands: NDP, BCN, MRD |

