Search Count: 2,237
 |
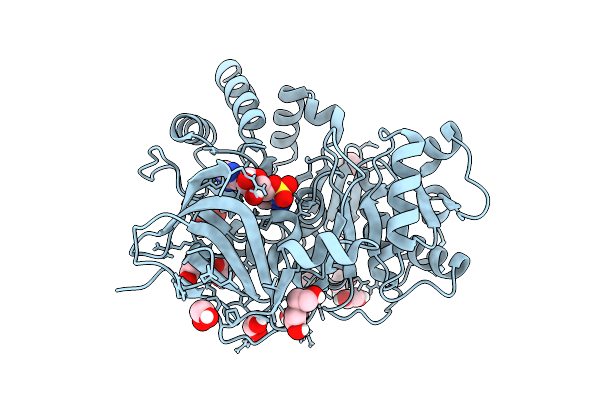
Cyro-Em Structure Of Prefusion Rsv Fusion Glycoprotein In Complex With Ziresovir And Motavizumab Fab
Organism: Human respiratory syncytial virus a2, Tequatrovirus t4, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.27 Å Release Date: 2026-01-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: A1EV1 |
 |
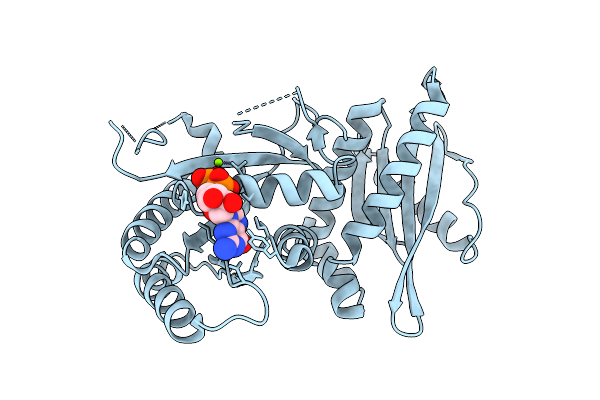
High-Resolution Crystal Structure Of Methyl-2,3-Diamino Propanoic Acid-Ams Inhibitor Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-12-10 Classification: LIGASE Ligands: A1BVA, EDO, PEG |
 |
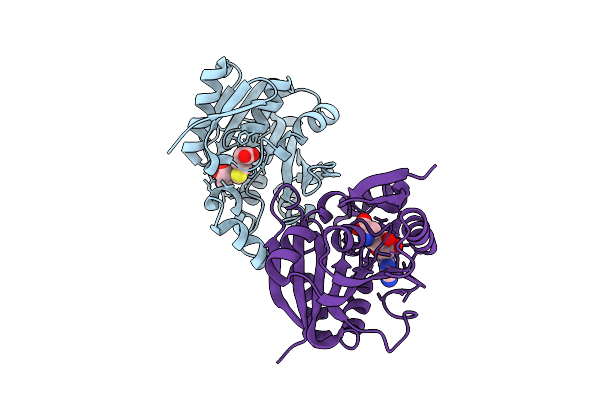
Crystal Structure Of Monomeric Rag-Like Small Gtpase From Asgard Lokiarchaeota (Lokiragm) In Complex With Gdp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GDP, MG |
 |
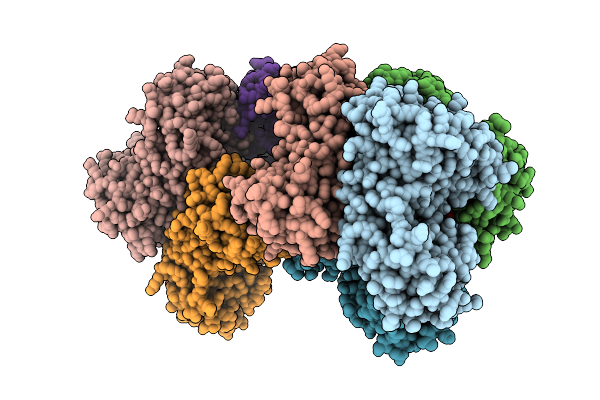
Crystal Structure Of The Carbamoyl N-Methyltransferase Asc-Orf2 Complexed With Sah
Organism: Amycolatopsis alba dsm 44262
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH |
 |
Crystal Structure Of Homo Tetrameric Raga-Like Small Gtpase From Asgard Lokiarchaeota (Lokirago) In Complex With Gtp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-09-10 Classification: HYDROLASE Ligands: GTP, MG |
 |
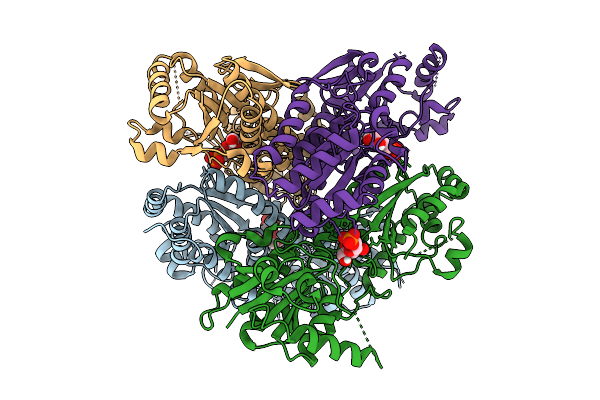
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-07-09 Classification: TRANSFERASE |
 |
Crystal Structure Of The Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Fructose 6-Phosphate
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: F6P |
 |
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Phosphoenolpyruvate, Phosphate And Mg2+
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: PEP, PO4, MG |
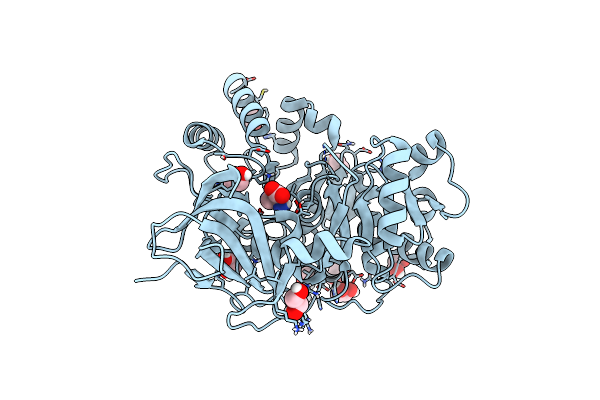 |
High-Resolution Crystal Structure Of 2,3-Diamino Propanoic Acid Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-05-28 Classification: LIGASE Ligands: EDO, DPP |
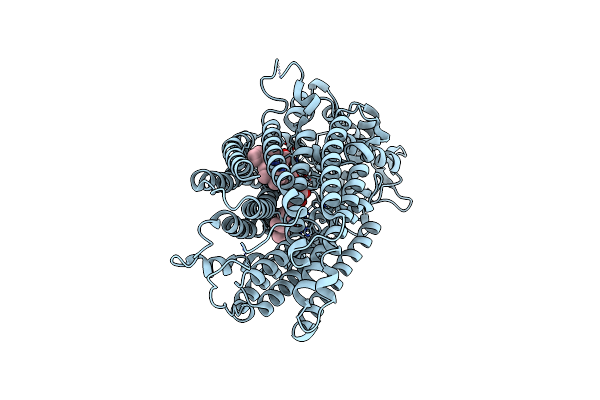 |
Cryoem Structure Of Monomeric Quinol Dependent Nitric Oxide Reductase From Neisseria Meningitidis
Organism: Neisseria meningitidis alpha14
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: HEM, FE, CA |
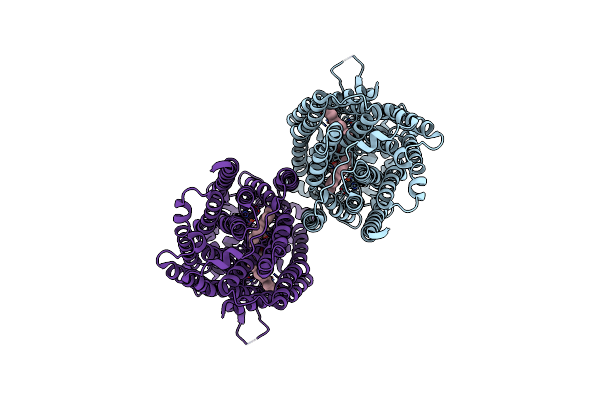 |
Cryoem Structure Of Dimeric Quinol Dependent Nitric Oxide Reductase From Neisseria Meningitidis
Organism: Neisseria meningitidis alpha14
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: HEM, FE, CA |
 |
Glycosyltransferase C From The Limosilactobacillus Reuteri Accessory Secretion System. Complex With Udp-Glcnac.
Organism: Limosilactobacillus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: UDP, CIT, GOL, EPE, CL, UD1 |
 |
Dimeric Structure Of The Longin Domain From A Lokiarchaeota Archaeon Roadblock-Longin Protein
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-03-26 Classification: UNKNOWN FUNCTION Ligands: MG |
 |
Roadblock Domain From A Lokiarchaeota Archaeon Roadblock-Longin Protein In P21
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2025-03-26 Classification: UNKNOWN FUNCTION |
 |
Roadblock Domain From A Lokiarchaeota Archaeon Roadblock-Longin Protein In P31
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-03-26 Classification: UNKNOWN FUNCTION Ligands: CL |
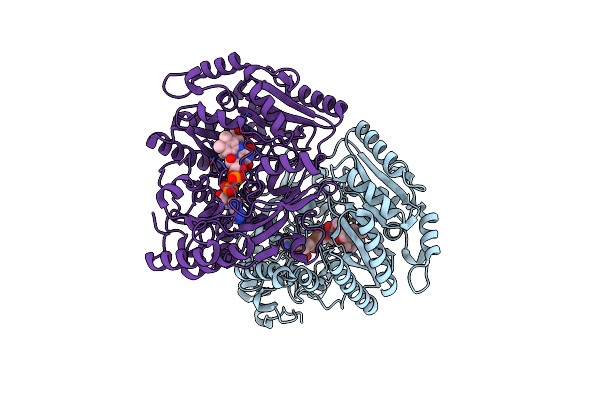 |
4-Allyl Syringol Oxidase From Streptomyces Cavernae: Complex With Propanol Syringol
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: QBF, FAD |
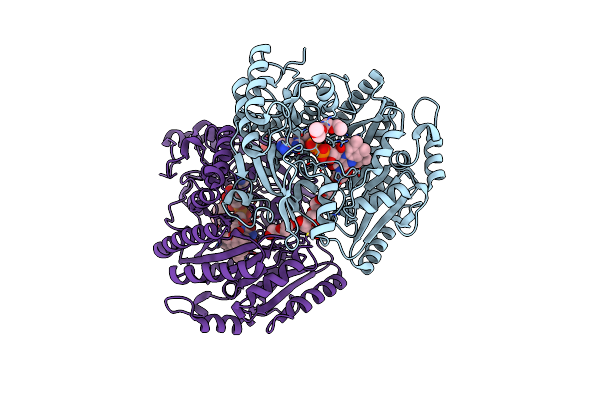 |
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: FAD, EOL, P6G, P4G, PEG |
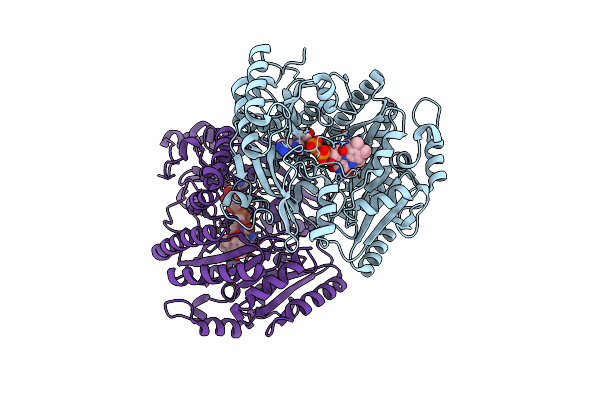 |
4-Allyl Syringol Oxidase From Streptomyces Cavernae: Complex With Vanillyl Alcohol
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: FAD, V55 |
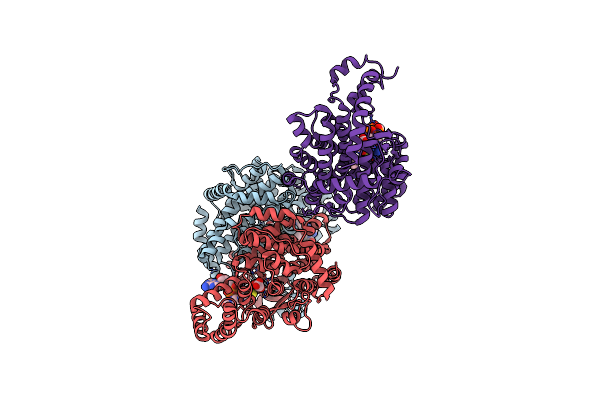 |
Organism: Thermosulfidibacter takaii
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: OAA, COA, ACE |
 |
Organism: Thermosulfidibacter takaii
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: OAA, ACO, COA |

