Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163-His280Ala With (S)-2-Fluoro-3-Phenylpropanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE |
 |
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163 - Lys181Lys/Trp185Tyr/His280Ala With (S)-2-Fluoro-3-(4-(Trifluoromethyl)Phenyl)Propanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1ES3 |
 |
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163 - Lys181Ser/Trp185Gly/His280Ala With (S)-2-Fluoro-3-(4-(Trifluoromethyl)Phenyl)Propanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1ES3 |
 |
Crystal Structure Of The Histidine Kinase Domain Of Bacteriophytochrome Rpbphp2
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2023-11-22 Classification: SIGNALING PROTEIN, TRANSFERASE Ligands: ATP, MG |
 |
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-08-16 Classification: FLUORESCENT PROTEIN Ligands: CYC |
 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound Phosphate
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: PO4, CA, NA |
 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound 2-Hydroxyquinoline
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: OCH, CA, NA |
 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound Tetrahedral Intermediate
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: CA, NA, SGU |
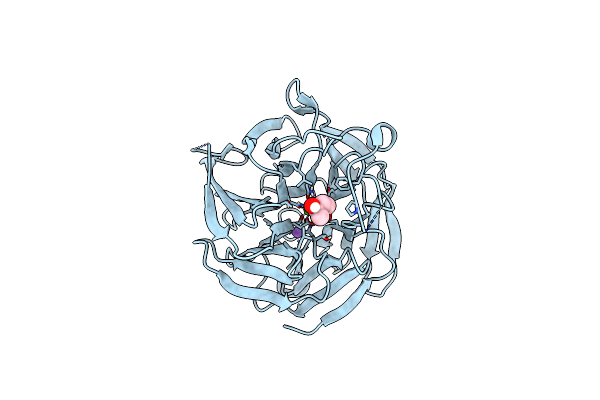 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound Product
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: CA, NA, SJ3 |
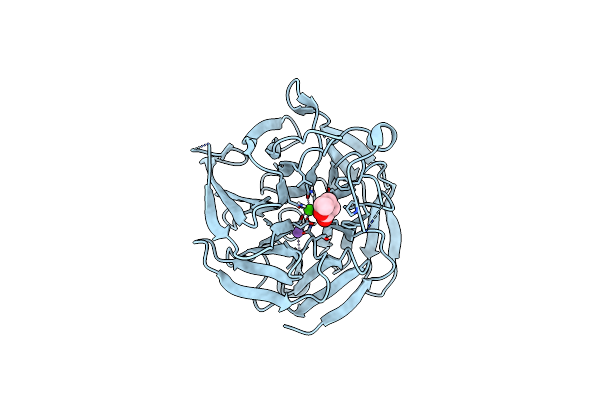 |
Crystal Structure Of Rpa3624, A Beta-Propeller Lactonase From Rhodopseudomonas Palustris, With Active-Site Bound (S)Gamma-Valerolactone
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: CA, NA, YVR |
 |
Solution Nmr Of The Specialized Apo-Acyl Carrier Protein (Rpa2022) From Rhodopseudomonas Palustris, Refined Without Rdcs. Northeast Structural Genomics Consortium Target Rpr324
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: SOLUTION NMR Release Date: 2022-08-10 Classification: TRANSFERASE |
 |
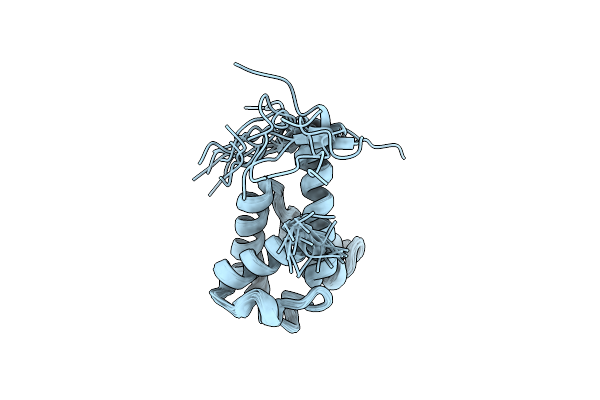
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-10-06 Classification: TRANSPORT PROTEIN Ligands: 4HP, EDO, PO4 |
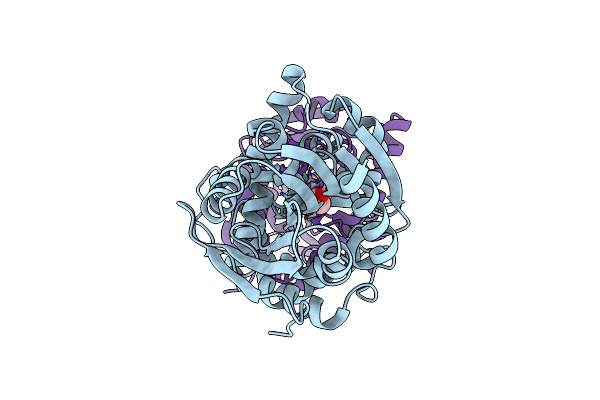 |
Time Resolved Structural Analysis Of The Full Turnover Of An Enzyme - 2256 Ms Covalent Intermediate 1
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009), Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-25 Classification: HYDROLASE Ligands: FAH |
 |
Time Resolved Structural Analysis Of The Full Turnover Of An Enzyme - 6788 Ms
Organism: Rhodopseudomonas palustris, Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-25 Classification: HYDROLASE Ligands: FAH |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 1 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using First 1 Degree Of Total 3 Degree Oscillation
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using Last 1 Degree Of Total 3 Degree Oscillation And 144 Kgy Dose
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Monomeric Crystal Structure Of Rpbphp1 Photosensory Core Domain From The Bacterium Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-02-06 Classification: PHOTOSYNTHESIS Ligands: BLA |
 |
Crystal Structure Of The Putative Class A Beta-Lactamase Penp From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-01-16 Classification: HYDROLASE Ligands: PG4, PEG, FMT, EDO, CL |