Search Count: 566
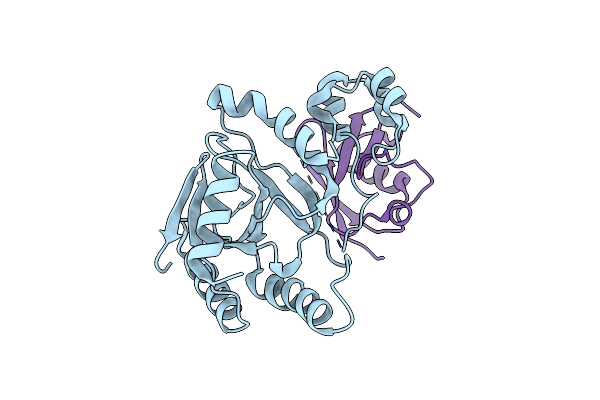 |
Crystal Structure Of A Semet-Labeled Effector From Chromobacterium Violaceum In Complex With Ubiquitin
Organism: Chromobacterium violaceum atcc 12472, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-22 Classification: TRANSFERASE |
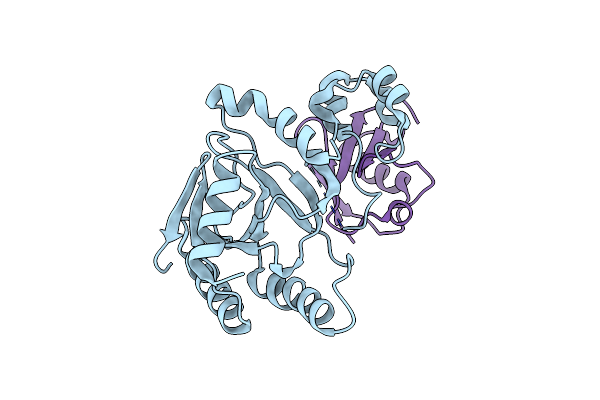 |
Crystal Structure Of An Effector From Chromobacterium Violaceum In Complex With Ubiquitin
Organism: Chromobacterium violaceum atcc 12472, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: TRANSFERASE |
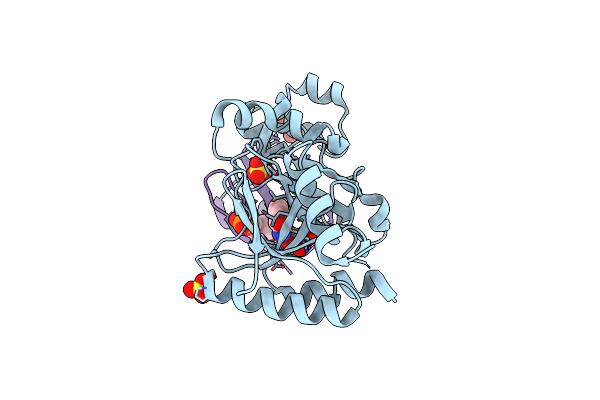 |
Organism: Chromobacterium violaceum atcc 12472, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SO4, GOL, NCA, AR6 |
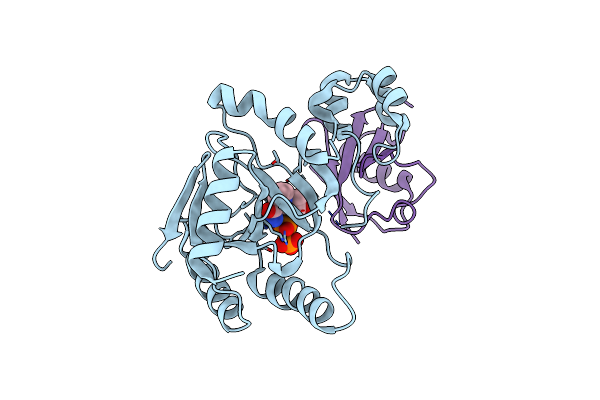 |
Organism: Chromobacterium violaceum atcc 12472, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: NAD |
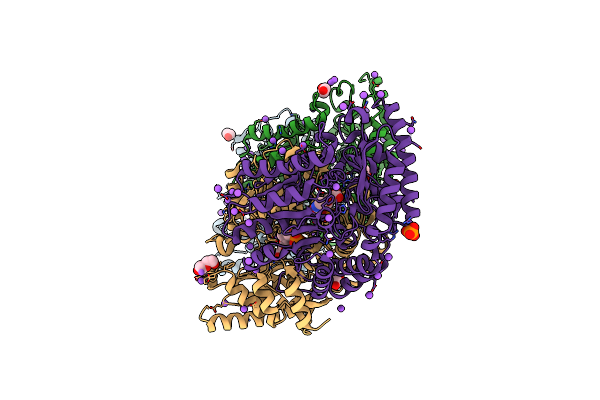 |
Crystal Structure Of Chromobacterium Violaceum Aminotransferase In Complex With Plp-Pyruvate Adduct
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: AN7, EDO, NA, PLP, PO4, PEG |
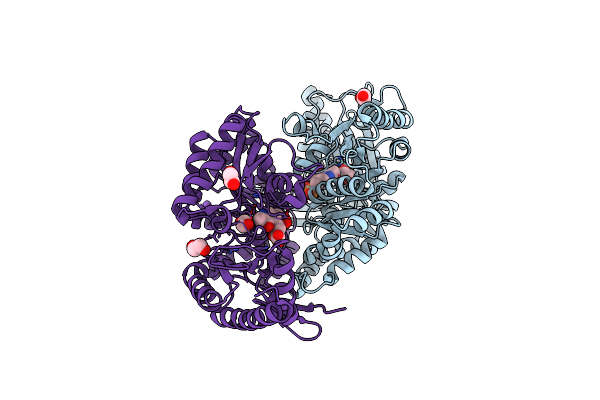 |
Crystal Structure Of Chromobacterium Violaceum Aminotransferase In Complex With Plp-Pyruvate Adduct
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: AN7, EDO, CL, NA |
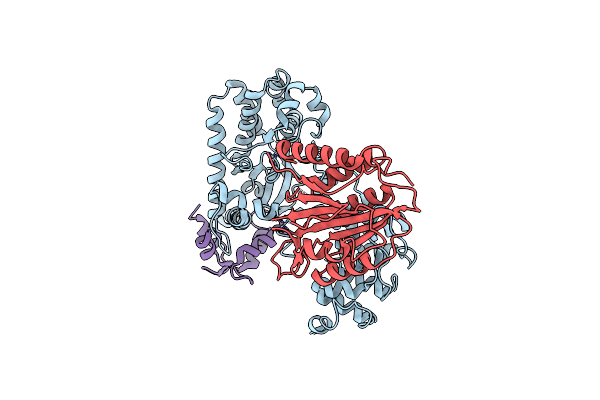 |
Crystal Structure Of Chromobacterium Violaceum Effector Copc In Complex With Host Calmodulin And Caspase-7
Organism: Chromobacterium violaceum atcc 12472, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2022-05-18 Classification: TRANSFERASE |
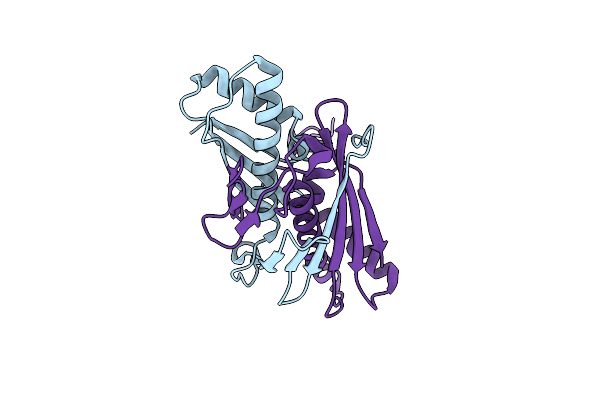 |
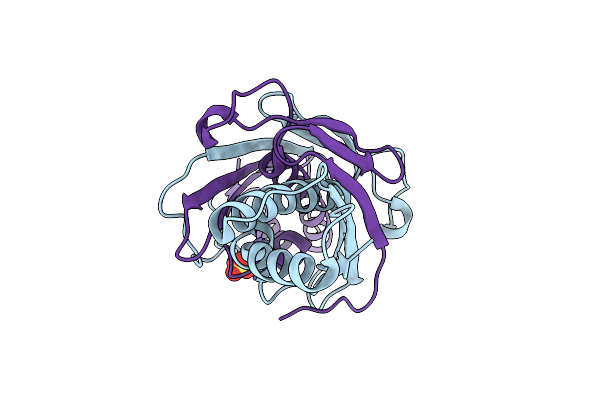
Ohra (Organic Hydroperoxide Resistance Protein) Wild Type From Chromobacterium Violaceum
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE |
 |
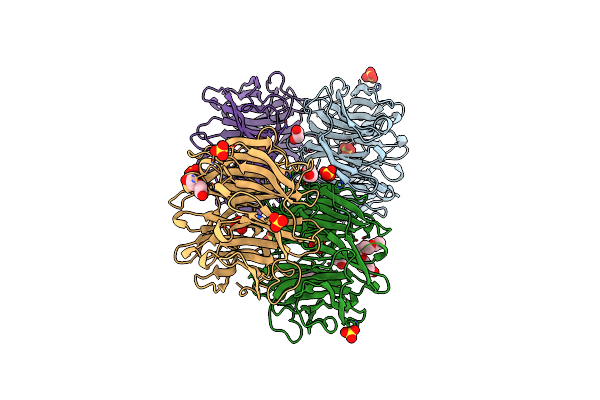
Ohra (Organic Hydroperoxide Resistance Protein) Mutant (C61S) In The "Open Conformation" From Chromobacterium Violaceum
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of Cv39L Lectin From Chromobacterium Violaceum At 1.8 A Resolution
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-12-04 Classification: SUGAR BINDING PROTEIN Ligands: SO4, EDO, PEG, 1PE |
 |
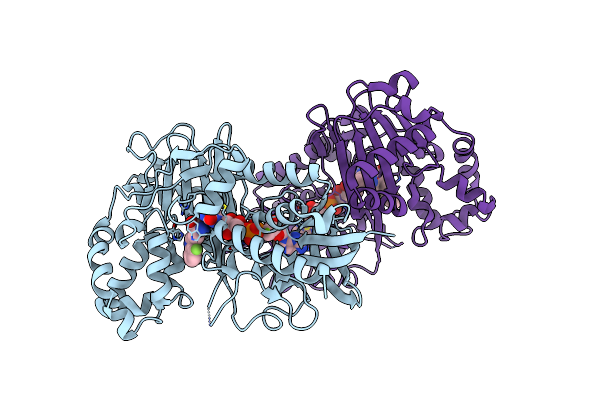
Crystal Structure Of The Omega Transaminase From Chromobacterium Violaceum In Complex With Pmp
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-07-17 Classification: TRANSFERASE Ligands: PMP, EDO, PEG |
 |
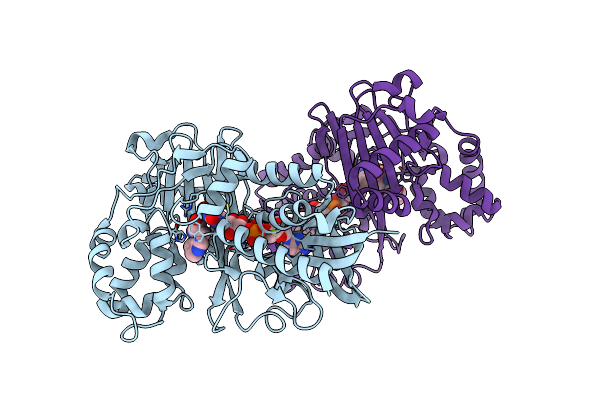
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 4-Fluoro-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, 4FW, MG |
 |
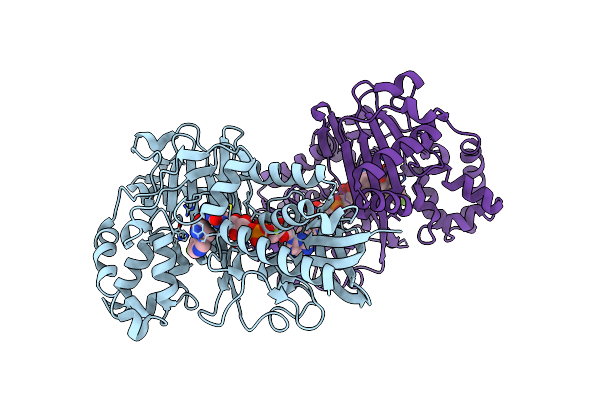
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 5-Methyl-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, D0Q, MG |
 |
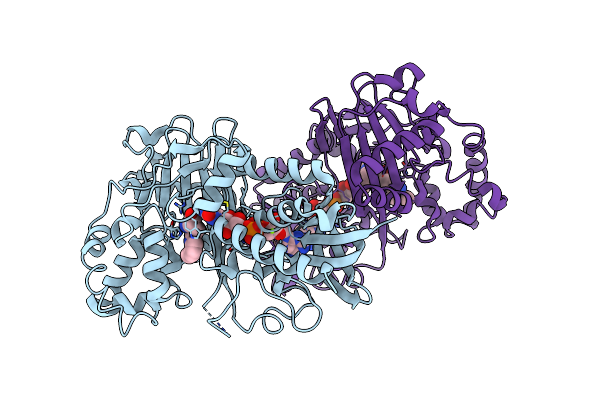
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 6-Fluoro-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, FT6, MG |
 |
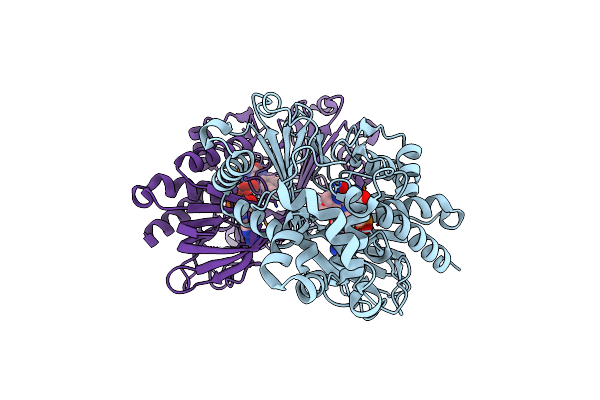
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 7-Methyl-L-Tryptophan
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, E95, MG |
 |
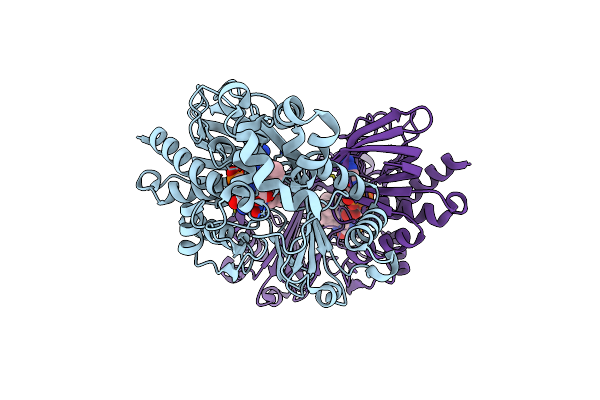
Crystal Structure Of Se-Met Tryptophan Oxidase (C395A Mutant) From Chromobacterium Violaceum
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
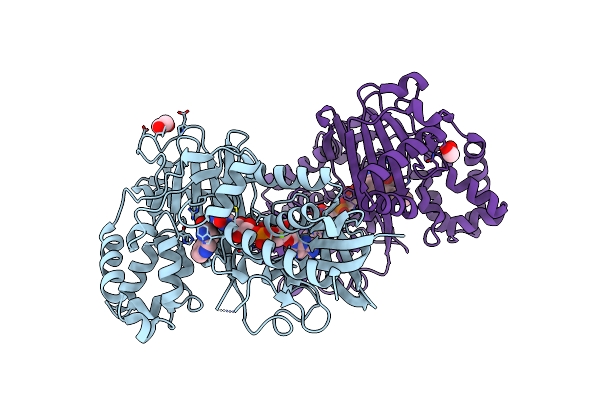
Crystal Structure Of Tryptophan Oxidase (C395A Mutant) From Chromobacterium Violaceum
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: TRP, FAD |
 |
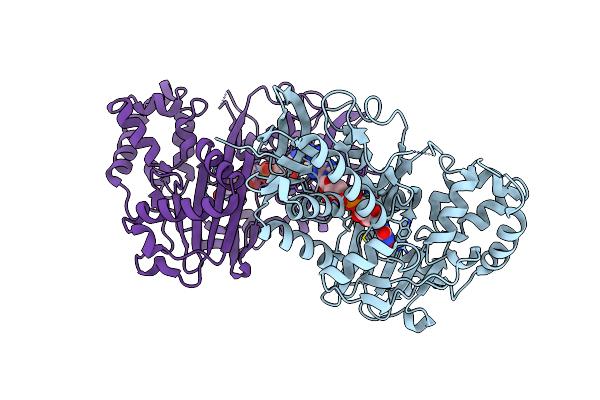
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-05-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, TRP, EDO, MG |
 |
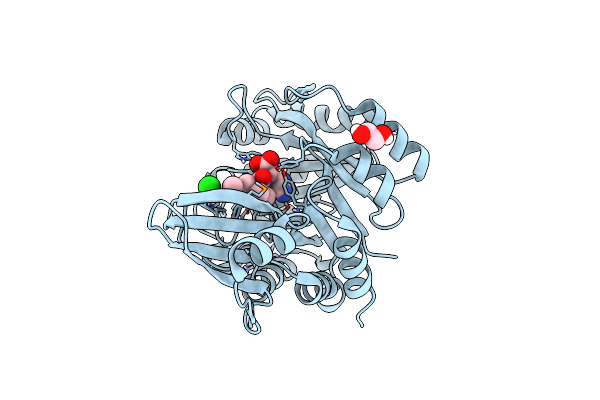
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-12-06 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, CL |
 |
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: EDO, CL, 6KY, HIS |

