Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
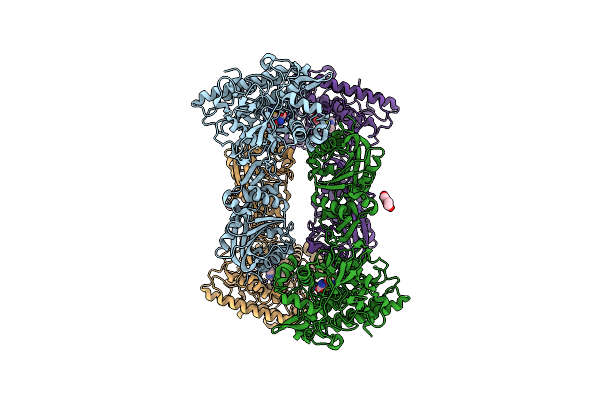 |
Bifunctional Sulfoxide Synthase Ovoa_Th2 In Complex With Histidine And Cysteine
Organism: Hydrogenimonas thermophila
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2023-12-06 Classification: OXIDOREDUCTASE Ligands: CO, HIS, CYS, PEG |
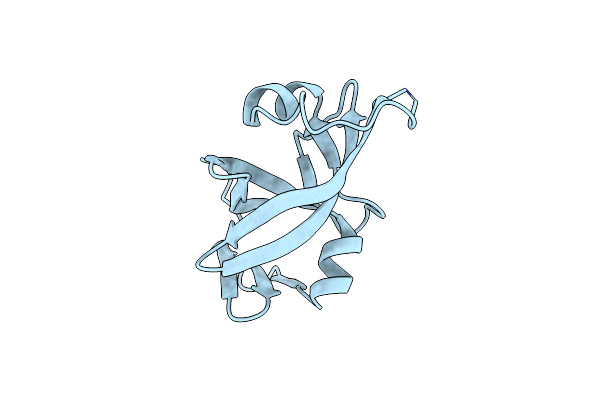 |
Crystal Structure Of Retroviral Protease-Like Domain Of Ddi1 From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-02-09 Classification: HYDROLASE |
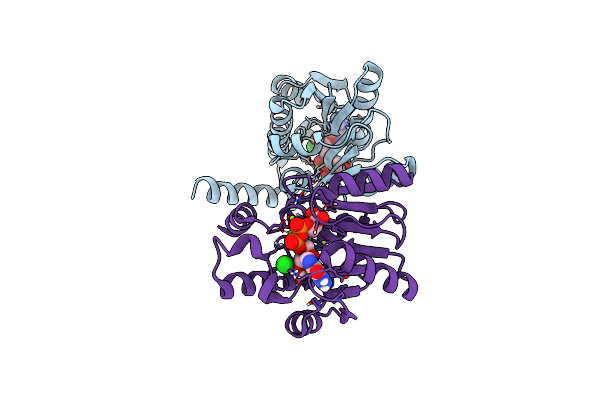 |
Structure Of 3-Phospho-D-Glycerate Guanylyltransferase With Product 3-Gppg Bound
Organism: Mycetohabitans rhizoxinica (strain dsm 19002 / cip 109453 / hki 454)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-02-02 Classification: TRANSFERASE Ligands: 6WI, MG, CL |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2021-10-20 Classification: ISOMERASE Ligands: MN, NA |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: GOL, MN |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: MPD, MN, RB0 |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MPD, MN, GOL |
 |
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma Mutant R148A
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma Mutant R148A In Complex With C8-Coa
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: CO8, TRS |
 |
Crystal Structure Of The Starter Condensation Domain Of The Rhizomide Synthetase Rzma Mutant H140V, R148A
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN |
 |
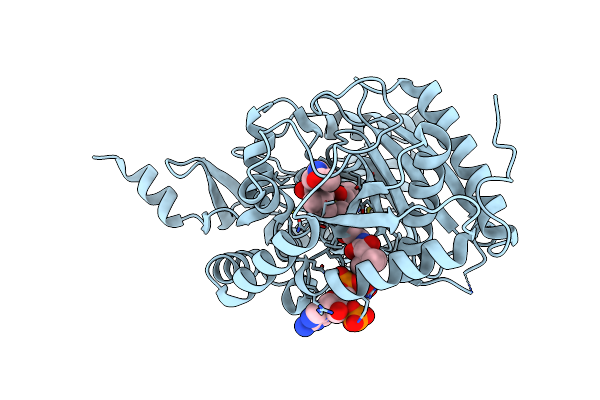
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma Mutant H140A/R148A In Complex With C8-Coa
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: CO8, TRS |
 |
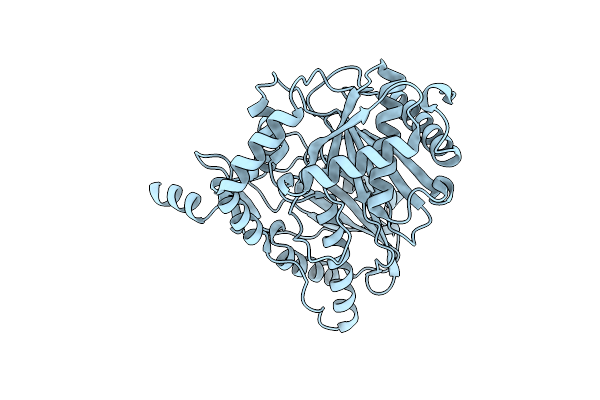
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma Mutant H140A/R148A In Complex With C8-Coa And Leu-Snac
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: CO8, FGU |
 |
Crystal Structure Of The Starter Condensation Domain Of Rhizomide Synthetase Rzma Mutant H140V/R148A In A "Product-Released" Conformation
Organism: Paraburkholderia rhizoxinica hki 454
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN |
 |
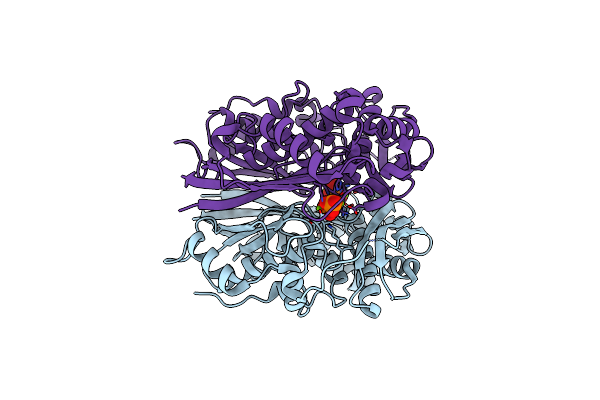
Crystal Structure Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis In Complex With Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: 3PO, MG, EU9 |
 |
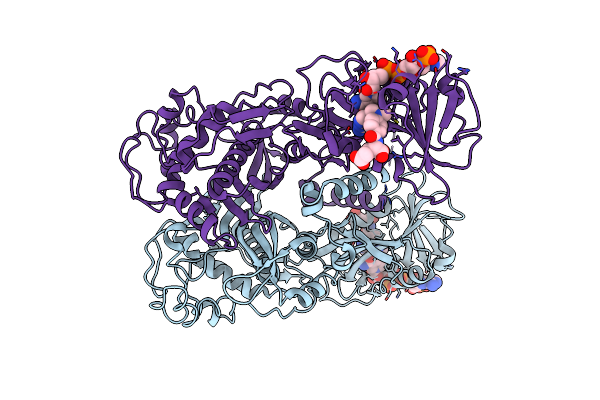
Crystal Structure Of Apo Form Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: MG, PO4 |
 |
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2020-06-17 Classification: TRANSFERASE Ligands: NDP, MTX |
 |
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-11-20 Classification: RNA BINDING PROTEIN Ligands: ZN, GOL, IPA, MG, PG4 |
 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)-4-Methoxyphenyl)Acetic Acid
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-10-16 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OED, MTX |
 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Benzoic Acid
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OG7, MTX, SO4 |