Search Count: 955
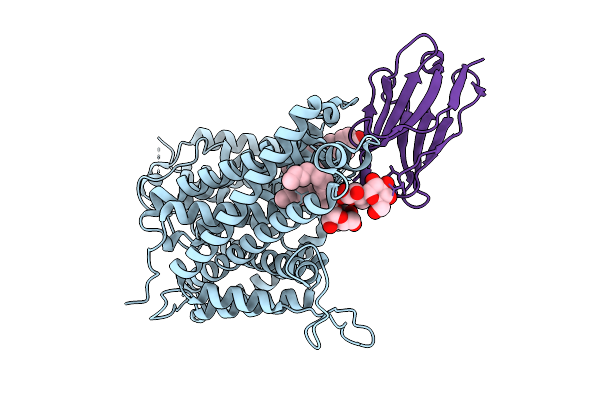 |
Cryo-Em Structure Of The Arabidopsis Thaliana Cat4 Transporter In The Outward-Open Apo State
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: CLR, AV0 |
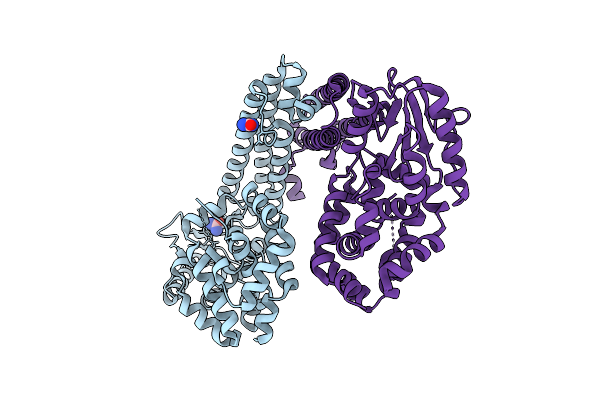 |
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
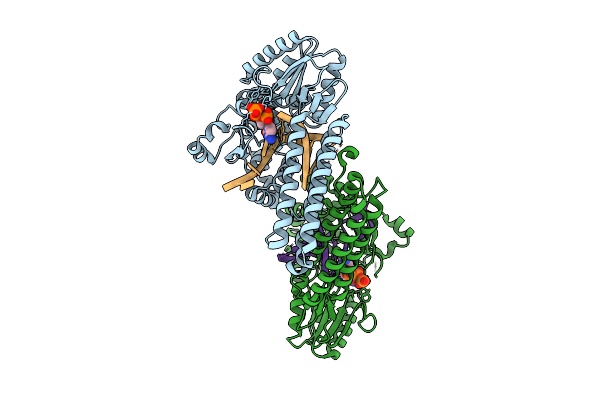 |
Organism: Gammaproteobacteria, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: DNA BINDING PROTEIN Ligands: MN, DCP |
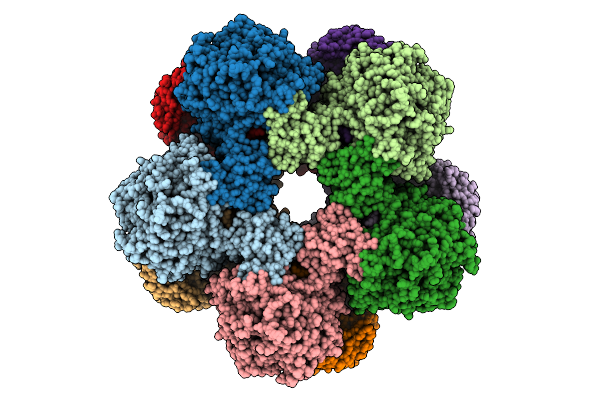 |
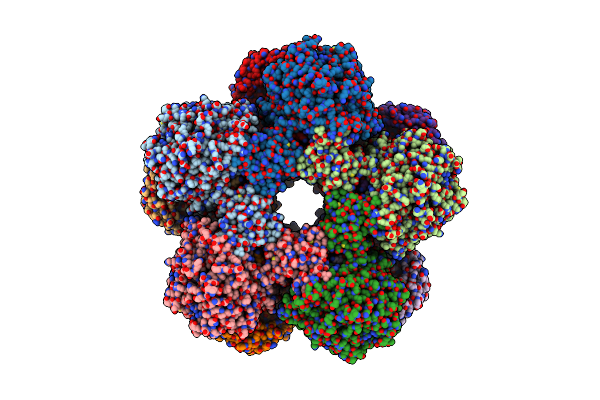
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei D-Hydrazino-Lysine Analog At 2.3 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LYASE Ligands: A1BD0 |
 |
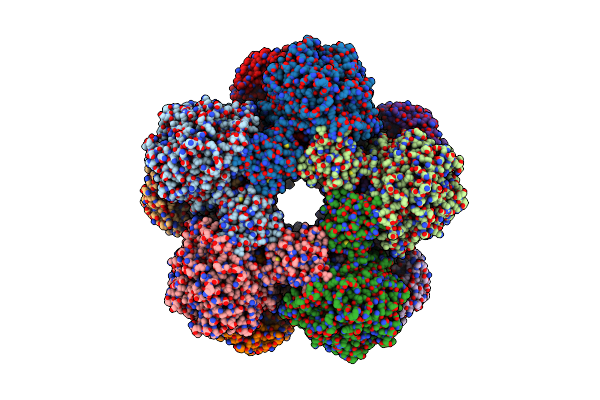
Cryoem Structure Of Holoenzyme Of Inducible Lysine Decarboxylase From Hafnia Alvei Holoenzyme At 2.19 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE |
 |
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei L-Hydrazino-Lysine Analog At 2.04 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE Ligands: A1BD1 |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN Ligands: 6WT |
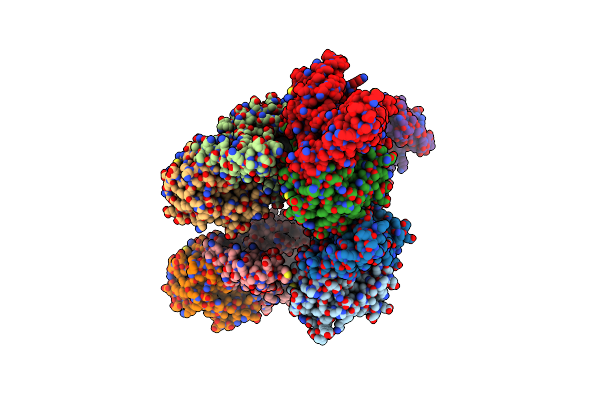 |
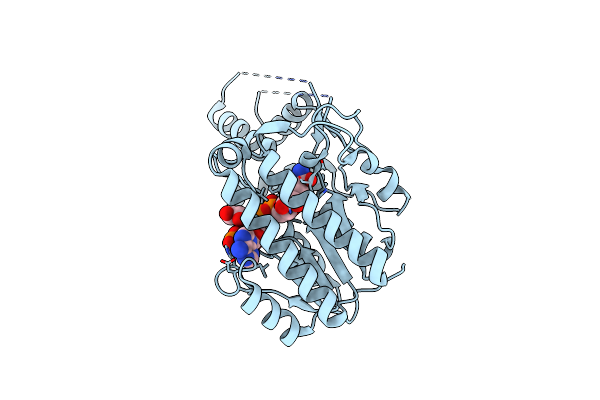
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanogenic Archaeon Iso4-G1
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-03-15 Classification: TRANSLATION |
 |
Organism: Brucella ovis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-10 Classification: ISOMERASE Ligands: NAP, GOL |
 |
Crystal Structure Of The Apo Enoyl-Acp-Reductase (Fabi) From Moraxella Catarrhalis
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With Nad And Triclosan
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAD, TCL, CA |
 |
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With The Cofactor Nad
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: NAD, CA, GOL |
 |
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-03-02 Classification: TRANSLATION Ligands: EDO, SO4 |
 |
Organism: Asticcacaulis sp. ybe204
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2021-11-10 Classification: UNKNOWN FUNCTION Ligands: ZN, MG |
 |
Crystal Structure Of Full-Length Streptococcal Bacteriophage Hyaluronidase In Complex With Unsaturated Hyaluronan Octa-Saccharides
Organism: Streptococcus pyogenes phage h4489a
Method: X-RAY DIFFRACTION Resolution:3.58 Å Release Date: 2021-06-09 Classification: LYASE |
 |
Crystal Structure Of Streptococcal Bacteriophage Hyaluronidase: Presence Of A Prokaryotic Collagen And Elucidation Of Catalytic Mechanism
Organism: Streptococcus pyogenes phage h4489a
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-05-12 Classification: LYASE Ligands: NI |

